Abstract
We present The Odonate Phenotypic Database (OPD): an online data resource of dragonfly and damselfly phenotypes (Insecta: Odonata). Odonata is a relatively small insect order that currently consists of about 6400 species belonging to 32 families. The database consists of multiple morphological, life-history and behavioral traits, and biogeographical information collected from literature sources. We see taxon-specific phenotypic databases from Odonata and other organismal groups as becoming an increasing valuable resource in comparative studies. Our database has phenotypic records for 1011 of all 6400 known odonate species. The database is accessible at http://www.odonatephenotypicdatabase.org/, and a static version with an information file about the variables in the database is archived at Dryad.
Measurement(s) | Invertebrate Taxonomy • body length • wing • appendage morphology trait • mating behavior • flight behavior • habitat • ecozone • climate • sexual dimorphism • size • color • biological pigment |
Technology Type(s) | digital curation |
Factor Type(s) | phenotypes |
Sample Characteristic - Organism | Zygoptera • Anisoptera |
Sample Characteristic - Environment | stream • pond • wetland ecosystem • lake • river • ephemeral spring |
Sample Characteristic - Location | South America • North America • Africa • Asia • Europe • Australia |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.10321595
Similar content being viewed by others
Background & Summary
The Odonate Phenotypic Database is an online data resource for dragonfly and damselfly phenotypes (Insecta: Odonata). The database consists of a variety of morphological, life-history and behavioral traits, and biogeographical information collected from various sources in the literature. The database is not intended for species identification, but for comparative analysis of insect groups or to be combined with data from other taxonomic groups. The database is provided along with a large phylogenetic tree (1322 taxa, 21% of known odonates https://www.pugetsound.edu/academics/academic-resources/slater-museum/biodiversity-resources/dragonflies/world-odonata-list2/, accessed in November 2015): “The Odonate Super Tree”. This phylogenetic tree was constructed using DNA-sequences from GenBank in combination with a traditional (morphologically-based) odonate taxonomy as our backbone1.
Comparative analyses are becoming an increasing common part of evolutionary studies, as researchers attempt to bridge the gap between microevolutionary processes and macroevolutionary patterns2,3,4,5,6,7,8. Most comparative analyses require on high-quality phenotypic data collected from the literature, and often a large amount of time is spent collecting such data. Trait information can be obtained from measurements of live and field-caught individuals, museum specimens or literature sources but often important covariates are missing, such as behavioural information or habitat data. It is therefore in the interest of many of us working in the field to collect and curate such phenotypic data in a coherent fashion so that such data can be used in future studies and combined with multiple other sources of phenotypic information, particularly in light of the explosion in phylogenetic comparative methods the last decades8,9.
Paradoxically, as a community we have been much more successful at storing and cataloguing genotypes, and DNA-sequences often exist in GenBank for many species, but not even basic phenotypic data (such as body size) exist in an easily accessible form for many organismal groups. This lack of information is most likely due to there being no clear structure or well-established routines agreed upon in how to store phenotypic data and which aspects of phenotypes should be measured. Phenotypic databases, because of the high-dimensional nature of most phenotypes, are also very different from a genetic database such as GenBank. Recent calls for a new field of “phenomics” – i.e. obtaining high-throughput phenotypic data in a similar fashion as in genomics – will always have to prioritize what aspects of the phenotypes to measure10,11. There are many challenges in developing such a general and cross-taxonomic research programme in phenomics, in particular the difficulties of establishing general and operational phenotype ontologies between distantly related organisms across the Tree of Life12,13.
From an evolutionary viewpoint, phenotypes are arguably just as important and interesting as genotypes, if not more so11,14,15, as selection operates on phenotypes, regardless of their genetic basis16. Increasingly integrative research practices in evolutionary biology will need not only access to high-quality genomic, molecular and phylogenetic resources, but will also need high-quality phenotypic and biogeographic data, fossil information for time-calibration of phylogenetic trees and other general data provided by biodiversity informatics17. The difficulty of these tasks and the size of each individual project should ideally not prevent the establishment of accessible structures needed to store the data.
One way forward is to create taxon-specific phenotypic databases, as we have done here. Having such databases available that focus on a certain taxonomic group, also allows the recorded phenotypes to be tailored to fit the needs of the specific group and have the advantages that trait definitions are less ambiguous. Examples of such open databases with various forms of phenotypic, biogeographic and phylogenetic data include AmphiBIO for amphibian ecological traits18, panTHERIA19 and EltonTraits 1.020 for birds and mammals, a global invasion atlas of birds21, Tree of Sex (a database on eukaryotic sex determination systems)22, a recent database on thermal developmental plasticity of reptiles23 and a global database on plants24. However, in the case of animals, such databases are largely focused on vertebrate groups, while the most speciose animal group – the insects – have few such open databases available. One exception is the Freshwater Biological Traits Database25 which contains trait data for North American macroinvertebrate taxa and includes habitat, life history, mobility, morphology, and ecological trait data, although not in all cases down to the species level.
We see taxon-specific phenotypic databases as becoming an increasingly valuable resource in comparative studies. Our research background and expertise is in the insect order Odonata (dragonflies and damselflies)(Fig. 1). To this end, we have collected data on 35 phenotypic, ecological and biogeographical variables that we see as useful to the research community (Online-only Table 1). Odonata is a relatively small insect order that currently consists of about 6400 species belonging to 32 families1. Odonata are morphologically highly conserved with respect to their overall external morphology (all species in this insect order have six legs and four wings), but they show considerable diversity in terms of wing and body colouration and shape (Fig. 1). Our database has at least some phenotypic records for 1011 of all 6400 known odonate species1 and a total of 3978 records (i.e., multiple records for many species, from different literature sources). The database is accessible at http://www.odonatephenotypicdatabase.org/.
Phenotypic and taxonomic diversity of Odonata. Phenotypic and taxonomic diversity of 12 representative families of Odonata, an insect order which currently encompasses c. a. 6400 species and a total of 32 families. All 32 odonate families are included in our molecular and time-calibrated phylogeny (Fig. 2). (a) Family Calopterygidae: Sapho orichalcea (Cameroon, Africa, January 2017). (b) Family Chlorocyphidae: Chlorocypha curta (Cameroon, Africa, January 2017). (c–d) Family Coenagrionidae: (c) Acanthagrion adustum (Guyana, South America, January 2015). (d) Argia moesta (Texas, North America, April 2012). (e) Family Lestidae: Lestes sponsa (Sweden, Europe, July 2010). (f) Family Synlestidae: Chlorolestes tessellatus (Eastern Cape, South Africa, Africa, April 2010). (g) Family Platycnemidae: Copera congolensis (Cameroon, Africa, February 2017). (h) Family Protoneuridae: Elattoneura balli (Cameroon, Africa, January 2017). (i–j) Family Aeshnidae: (i) Aeshna affinis (Sweden, Europe, August 2010). (j) Anax imperator (Sweden, Europe, August 2015). (k) Family Cordulegasteridae: Cordulegaster boltonii (Sweden, Europe, July 2016). (l) Family Corduliidae: Somatochlora metallica (Sweden, Europe, June 2014). (m) Family Libellulidae: Zenithoptera fasciata (Guyana, South America, January 2015). (n) Family Gomphidae: Ictinogomphus ferox (Namibia, Africa, April 2017). All photographs by Erik Svensson.
Methods
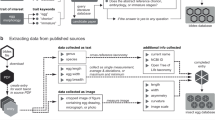
We have collected phenotypic data and data on habitats, climatic classifications and more coarse-grained biogeographic categories (e. g. ecozones26), from the scientific literature and from odonate field guides (Online-only Table 1). The field guides from which we obtained our phenotypic data are listed in Online-only Table 2. Phenotypes were scored by following a specific set of instructions for each variable that are described in an accompanying file that is uploaded alongside this paper as Supporting Material (“VariableDefinitions.pdf”) which is available on the Dryad Digital Repository26. The descriptions of each variable can be found in the Data Records section below and in Online-only Table 1. The construction of the Odonate Phylogenetic Super Tree has been described in detail elsewhere1.
Developing general and meaningful phenotype ontologies that are generally applicable across many taxa is a challenging task that is beyond the scope of this paper. The phenotypic variables (size, behaviours, wing and body colouration, colour patterns; Online-only Table 1) that we have assembled from the literature are not always easily and straightforward to translate to other, more well-studied insect groups, including the classical model organism Drosophila melanogaster, although our classifications are largely consistent with the recommendations given in the Ontobee database (http://www.ontobee.org/) and the Phenotype and Trait Ontology (PATO) database (http://www.ontobee.org/ontology/PATO)(Online-only Table 1). Clearly a lot of work is needed before we can develop generally applicable phenotypic ontologies across the entire class Insecta, let alone across more distantly related organismal groups12,13. All of our size measurements are given in millimetres (mm), although we acknowledge the fact that even a simple variable like wing length can be measured in different ways in different studies.
With respect to the ecological and geographic variables, our database is more easily connected and comparable with similar initiatives from other organismal groups. The ecozone variable for the distribution data of the different species follows the recent updates and classifications of Wallace’s classical zoogeographic regions by Holt et al.27. Our habitat classifications of water bodies (Online-only Table 1) are largely consistent with the environmental ontologies given in the Environment Ontology database (ENVO: http://www.ontobee.org/ontology/ENVO). We define lakes, ponds, rivers and streams in the same way as ENVO, whereas our classification of Ephemeral wetlands encompass both ephemeral springs (http://www.ontobee.org/ontology/ENVO?iri=http://purl.obolibrary.org/obo/ENVO_00000204) and ephemeral rivers (http://www.ontobee.org/ontology/ENVO?iri=http://purl.obolibrary.org/obo/ENVO_01000979).
Data Records
The database has been deposited on the Dryad Digital Respository26, and additional material, including the code that is also uploaded on Github (see above) is also deposited with the most recent and updated database version at http://www.odonatephenotypicdatabase.org/. This data includes a PDF-file that describes each of the variables within the database and how they were collected (“VariableDefinitions.pdf”)26.
We note that our database contains information from only 1011 of all 6400 species, i. e. about 16% taxonomic coverage, and coverage varies among traits (Fig. 2). For instance, behavioural traits like male guarding of females and territoriality have considerably lower coverage than morphological traits like size and other phenotypic measurements (Online-only Table 1; Fig. 2). The reason for this is that morphological traits can easily be obtained also from dead specimens in museums, whereas behavioural traits would typically require time-consuming field studies of these insects28,29, particularly in the tropics where species diversity is high but where we still lack basic faunistic information even about which species occur in many countries30. It is particularly noteworthy that much data overall is lacking from several tropical damselfly families (Calopterygidae, Chlorocyphidae, Euphalidae, Megapodagrionidae, Polythoridae and Platystictidae) and some dragonfly families, particularly Gomphidae (Fig. 2). Our dataset illustrated in Fig. 2 should hopefully serve as a basis for targeted studies on these families.
Finally, we also note that information about the occurrence of female colour polymorphisms, which is an important feature of many odonate species and which have important roles in frequency-dependent sexual conflict31,32,33,34,35,36,37 is missing for many species and genera. Documenting the presence or absence of female colour polymorphism typically requires large sample sizes, particularly if one wishes to estimate the frequencies of certain morphs, such as male-mimicking females, and such information is typically available for mainly temperate species, such as those in the genera Ischnura and Enallagma35,38. There is clearly a need for more quantitative field studies and surveys to improve this situation and fill the missing gaps in our knowledge, particularly for tropical taxa.
Technical Validation
All of the phenotypic data were collected from published field guides or reliable internet sources. The field guides are listed in Online-only Table 2. All the field guides have been published by respected odonatologists and experts on species identification. Our database is therefore not static, and additional data will be added as it becomes available. Researchers interested in contributing to this project are encouraged to contact the author for correspondence on how to incorporate new data. We will accept data both from already published sources (e. g. scientific papers) even if it has already been deposited in other databases such as Dryad, as well as data that is not intended to be published elsewhere, as long as it can be tailored to the format of the Odonate Phenotypic Database. Each species has a reference list, which lists the references from where the data were gathered, so it is possible to check each entry against these primary sources.
Usage Notes
The database is intended to be used in future comparative analyses of odonate trait evolution. We therefore provide a previously published phylogenetic tree (Fig. 2) along with these phenotypic data. Previous questions that we have addressed in recent past using phylogenetic comparative methods and these and similar data include the relationship between latitude and wing pigmentation39 and the micro- and macroevolutionary dynamics of body size1. We have also investigated diversification dynamics (speciation and extinction rates) in relation to body size and wing pigmentation1,39. Body size is also a trait that is of interest for conservation biology, as extinction risk (as defined by IUCN redlist criteria) was recently demonstrated to be significantly related to body size in damselflies40. In addition, behavioural diversity (“ethodiversity”) is a neglected aspect of biodiversity and conservation biology29 and in the future, we hope to incorporate such information in this database.
Other interesting questions to pursue in even more depth in the future include the evolutionary and ecological dynamics of sexual conflict and its consequences for the maintenance of sex-limited colour polymorphism in females31,32,33,34,35,36, and how climatic niche conservatism41,42 can shape the evolution, ecology and biogeography of odonate range limits43,44,45,46.
Code availability
All code used to generate the website and Odonate Super Tree are available on Github (https://github.com/jhnwllr/shiny-server/tree/master/odonates).
Change history
06 February 2020
The ‘Acknowledgements’ section was updated to recognize open access funding support.
References
Waller, J. T. & Svensson, E. I. Body size evolution in an old insect order: No evidence for Cope’s Rule in spite of fitness benefits of large size. Evolution 71, 2178–2193 (2017).
Arnold, S. J., Pfrender, M. E. & Jones, A. G. The adaptive landscape as a conceptual bridge between micro- and macroevolution. Genetica 112–113, 9–32 (2001).
Arnold, S. J. Phenotypic evolution: the ongoing synthesis. Am. Nat. 183, 729–746 (2014).
Estes, S. & Arnold, S. J. Resolving the paradox of stasis: Models with stabilizing selection explain evolutionary divergence on all timescales. Am.Nat. 169, 227–244 (2007).
Uyeda, J. C., Hansen, T. F., Arnold, S. J. & Pienaar, J. The million-year wait for macroevolutionary bursts. Proc. Natl. Acad. of Sci., USA 108, 15908–15913 (2011).
Hansen, T. F. Adaptive Landscapes and macroevolutionary dynamics. In The Adaptive Landscape in Evolutionary Biology. (eds. Svensson, E. I. & Calsbeek, R.) (Oxford University Press, 2012).
Svensson, E. I. & Calsbeek, R. The Adaptive Landscape in Evolutionary Biology. (Oxford University Press, 2012).
Garamszegi, L. Z. Modern phylogenetic comparative methods and their application in evolutionary biology: concepts and practice. (Springer, 2014).
Pennell, M. W. & Harmon, L. J. An integrative view of phylogenetic comparative methods: connections to population genetics, community ecology, and paleobiology. In Year in Evolutionary Biology. (ed.s Mousseau, T. A. & Fox, C. W.) vol. 1289, 90–105 (Blackwell Science Publ, 2013).
Houle, D. Numbering the hairs on our heads: The shared challenge and promise of phenomics. Proc. Natl. Acad. of Sci., USA 107, 1793–1799 (2010).
Houle, D., Govindaraju, D. R. & Omholt, S. Phenomics: the next challenge. Nature Rev. Gen. 11, 855–866 (2010).
Burleigh, J. G. et al. Next-generation phenomics for the Tree of Life. PLoS Curr. 5 (2013).
Deans, A. R. et al. Finding our way through phenotypes. PLOS Biol. 13, e1002033 (2015).
Kühl, H. S. & Burghardt, T. Animal biometrics: quantifying and detecting phenotypic appearance. Trends Ecol. Evol. 28, 432–441 (2013).
Laughlin, D. C. & Messier, J. Fitness of multidimensional phenotypes in dynamic adaptive landscapes. Trends Ecol. Evol. 30, 487–496 (2015).
Lande, R. & Arnold, S. J. The measurement of selection on correlated characters. Evolution 37, 1210–1226 (1983).
Losos, J. B. et al. Evolutionary biology for the 21st century. PLoS. Biol. 11, e1001466 (2013).
Oliveira, B. F., São-Pedro, V. A., Santos-Barrera, G., Penone, C. & Costa, G. C. AmphiBIO, a global database for amphibian ecological traits. Sci. Data 4, 170123 (2017).
Jones, K. E. et al. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648–2648 (2009).
Wilman, H. et al. EltonTraits 1.0: Species-level foraging attributes of the world’s birds and mammals. Ecology 95, 2027–2027 (2014).
Dyer, E. E., Redding, D. W. & Blackburn, T. M. The global avian invasions atlas, a database of alien bird distributions worldwide. Sci. Data 4, 170041 (2017).
The Tree of Sex Consortium. et al. Tree of Sex: A database of sexual systems. Sci. Data 1, 140015 (2014).
Noble, D. W. A. et al. A comprehensive database of thermal developmental plasticity in reptiles. Sci. Data 5, 180138 (2018).
Kattge, J. et al. TRY – a global database of plant traits. Global Change Biol. 17, 2905–2935 (2011).
Environmental Protection Agency. U. S. Freshwater Biological Database (Traits), https://www.epa.gov/risk/freshwater-biological-traits-database-traits#tab-2 (2019).
Waller, J. T., Willink, B., Tschol, M. & Svensson, E. I. Data from: The odonate phenotypic database, a new open data resource for comparative studies of an old insect order. Dryad Digital Repository, https://doi.org/10.5061/dryad.15pm5qc (2019).
Holt, B. G. et al. An Update of Wallace’s Zoogeographic Regions of the World. Science 339, 74–78 (2013).
Svensson, E. I., Eroukhmanoff, F., Karlsson, K., Runemark, A. & Brodin, A. A role for learning in population divergence of mate preferences. Evolution 64, 3101–3113 (2010).
Cordero-Rivera, A. Behavioral Diversity (Ethodiversity): A Neglected Level in the Study of Biodiversity. Front. Ecol. Evol. 5 (2017).
Ellenrieder, N., von Castro, B. W. & Svensson, E. Checklist of the dragonflies and damselflies from Guyana (Insecta: Odonata), with new records from the country. Check List 13(2), 1–22 (2017).
Svensson, E. I., Abbott, J. & Hardling, R. Female polymorphism, frequency dependence, and rapid evolutionary dynamics in natural populations. Am. Nat. 165, 567–576 (2005).
Gosden, T. P. & Svensson, E. I. Density-dependent male mating harassment, female resistance, and male mimicry. Am. Nat. 173, 709–721 (2009).
Takahashi, Y., Yoshimura, J., Morita, S. & Watanabe, M. Negative frequency-dependent selection in female color polymorphism of a damselfly. Evolution 64, 3620–3628 (2010).
Takahashi, Y., Kagawa, K., Svensson, E. I. & Kawata, M. Evolution of increased phenotypic diversity enhances population performance by reducing sexual harassment in damselflies. Nature. Comm. 5, 4468 (2014).
Le Rouzic, A., Hansen, T. F., Gosden, T. P. & Svensson, E. I. Evolutionary time-series analysis reveals the signature of frequency-dependent selection on a female mating polymorphism. Am. Nat. 185E, E182–E196 (2015).
Gering, E. J. Male-mimicking females increase male-male interactions, and decrease male survival and condition in a female-polymorphic damselfly. Evolution 71, 1390–1396 (2017).
Willink, B., Duryea, M. C. & Svensson, E. I. Macroevolutionary origin and adaptive function of a polymorphic female signal involved in sexual conflict. Am. Nat. 194, 707–724 (2019).
Fincke, O. M., Jödicke, R., Paulson, D. & Schultz, D. T. The evolution and frequency of female color morphs in Holarctic Odonata: why are male-like morphs typically the minority? Int. J. Odonatol. 8, 183–212 (2005).
Svensson, E. I. & Waller, J. T. Ecology and sexual selection: evolution of wing pigmentation in calopterygid damselflies in relation to latitude, sexual dimorphism and speciation. Am. Nat. 182, E174–E195 (2013).
Suárez-Tovar, C. M., Rocha-Ortega, M., González-Voyer, A., González-Tokman, D. & Córdoba-Aguilar, A. The larger the damselfly, the more likely to be threatened: a sexual selection approach. J. Insect Conserv. 23, 535–545 (2019).
Wiens, J. J. & Graham, C. H. Niche Conservatism: Integrating Evolution, Ecology, and Conservation Biology. Ann. Rev. Ecol. Evol. Syst. 36, 519–539 (2005).
Svensson, E. I. Non-ecological speciation, niche conservatism and thermal adaptation: how are they connected? Org., Div. Evol. 12, 229–240 (2012).
Wellenreuther, M., Larson, K. W. & Svensson, E. I. Climatic niche divergence or conservatism? Environmental niches and range limits in ecologically similar damselflies. Ecology 93, 1353–1366 (2012).
Lancaster, L. T., Dudaniec, R. Y., Hansson, B. & Svensson, E. I. Latitudinal shift in thermal niche breadth results from thermal release during a climate-mediated range expansion. J. Biogeography 42, 1953–1963 (2015).
Lancaster, L. T. et al. Gene expression under thermal stress varies across a geographic range expansion front. Mol. Ecol. 25, 1141–1156 (2016).
Dudaniec, R. Y., Yong, C. J., Lancaster, L. T., Svensson, E. I. & Hansson, B. Signatures of local adaptation along environmental gradients in a range-expanding damselfly (Ischnura elegans). Mol. Ecol. 27, 2576–2593 (2018).
Theischinger, G. & Hawking, J. The Complete Field Guide to Dragonflies of Australia. (Csiro Publishing, 2006).
Ozono, A. & Futahashi, R. Dragonflies of Japan. (unichi-Sogo Syuppan, 2012).
Dijkstra, K.-D. B. & Lewington, R. Field Guide to the Dragonflies of Britain and Europe. (British Wildlife Publishing, 2006).
Paulson, D. Dragonflies and Damselflies of the West. (Princeton University Press, 2009).
Samways, M. J. Dragonflies and damselflies of South Africa. (Pensoft, 2008).
Tarboton, W. & Tarboton, M. A fieldguide to the Dragonflies of South Africa. (Privately published by the authors, 2002).
Heckman, C. W. Encyclopedia of South American Aquatic Insects: Odonata-Anisoptera. (Springer, 2006).
Paulson, D. Dragonflies and Damselflies of the East. (Princeton University Press, NJ, 2012).
Bun, T. H., Keng, W. L. & Hämäläinen, M. A Photographic Guide to the Dragonflies of Singapore. (Lee Kong Chian Natural History Museum, 2010).
Subramanian, K. A. Dragonflies and Damselflies of Peninsular India: A Field Guide. (Centre for Ecological Sciences, Indian Institue of Science, https://www.ias.ac.in/public/Resources/Other_Publications/Overview/Dragonflies/odonates_introduction.pdf, 2005).
Hämäläinen, M. & Pinratana, A. Atlas of the Dragonflies of Thailand Distribution Maps by Provinces. (Brothers of St Gabriel in Thailand, 1999).
Herrara, C. E. Dragonflies and damselflies of Middle America and the Caribbean. (INBio: the Costa Rican Biodiversity Institute, 2006).
Garrison, R. W., Von Ellenrieder, N. & Louton, J. A. Dragonfly Genera of the New World: An Illustrated and Annotated Key to the Anisoptera. (Johns Hopkins University Press, 2006).
Nair, M. V. Dragonflies & Damselflies of Orissa and Eastern India. (Orissa Wildlife Organisation, 2011).
Tarboton, M. & Tarboton, W. A fieldguide to the Damselflies of South Africa. (Privately published by the authors, 2005).
Sugimura, M., Ishida, S., Kojima, K., Ishida, K. & Aoki, T. Dragonflies of the Japanese archipelago in colour. (Hokkaido University Press, 2001).
Lencioni, F. A. A. Damselflies of Brazil: an illustrated identification guide. Volumes 1 and 2. (F.A.A Lencioni, 2006).
Suhling, F. & Martens, A. Dragonflies and Damselflies of Namibia. (Gamsberg Macmillan, 2007).
Askew, R. R. The dragonflies of Europe. (B. H. & A. Harley Ltd, 1988).
Dunkle, S. W. Dragonflies through Binoculars: A Field Guide to Dragonflies of North America. (Oxford University Press, 2000).
Manolis, T. D. Dragonflies and Damselflies of California. (University of California Press, 2003).
Abbott, J. C. Damselflies of Texas: A Field Guide. (University of Texas Press, 2011).
Dijkstra, K.-D. B. & Clausnitzer, V. The Dragonflies and Dameselflies of Eastern Africa: Handbook for All Odonata from Sudan to Zimbabwe. (Koninklijk Museum Voor Midden-Afrika, 2014).
Tze-Wai, T. et al. The Dragonflies of Hong Kong. (Huayu Nature Book Trade Co.Ltd, 2011).
Michalski, J. The Dragonflies and Damselflies of Trinidad and Tobago. (Kanduanum Books, 2015).
Kompier, T. A Guide to the Dragonflies and Damselflies of the Serra dos Orgaos, South-Eastern Brazil. (REGUA Publications, 2015).
Marinov, M. & Waqa-Sakiti, H. An Illustrated Guide to Dragonflies of Viti Levu, Fiji. (University of the South Pacific Press, 2013).
Polhemus, D. & Asquith, A. Hawaiian Damselflies: A Field Identification Guide. (Bishop Museum Pr; trade pbk edition, 1996).
Biggs, K. Common Dragonflies of California. (Azalea Creek Publishing, 2000).
Garrison, R. W., Von Ellenrieder, N. & Louton, J. A. Damselfly Genera of the New World: An Illustrated and Annotated Key to the Zygoptera. (Johns Hopkins University Press, 2010).
Acknowledgements
We would like to acknowledge the students and laboratory assistants that helped us to collect the data from the literature, and in particular Anna Kell, Ev Poslin, Hanna Bensch, Robin Pranter, Kajsa Svensson, Lisa Winberg, Karolina Pehrson, Mireia Balesta and Tammy Ho. We would like to also thank the many authors of the field guides from which our database references. Our database is by no means nor intends to be a substitute for these valuable books. Funding for this study have been provided by research grants from The Swedish Research Council (VR: grant no. 2016-03356), Gyllenstiernska Krapperupstiftelsen (grant no. KR2018-0038) and Olle Engqvist Byggmästares Stiftelse to E.I.S. and from a Faculty Mobility grant from the University of Costa Rica and a grant from the Schlumberger Foundation to B.W. Open access funding provided by Lund University.
Author information
Authors and Affiliations
Contributions
John Waller: constructed the website as part of his PhD-work, collected data, helped create variable definitions, wrote the first draft of the manuscript. Beatriz Willink: collected data, helped plan and organize data collection. Maximilian Tschol: collected wing pigment data, created variable definitions. Erik I. Svensson: created variable definitions, supervised data collection, came up with the original idea to create the database and secured funding for the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online-only Tables
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Waller, J.T., Willink, B., Tschol, M. et al. The odonate phenotypic database, a new open data resource for comparative studies of an old insect order. Sci Data 6, 316 (2019). https://doi.org/10.1038/s41597-019-0318-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-019-0318-9
This article is cited by
-
From the forest to the city: the persistence of dragonflies and damselflies in the urban jungle
Biodiversity and Conservation (2024)
-
Wolbachia-driven selective sweep in a range expanding insect species
BMC Ecology and Evolution (2021)