Abstract
Dioecy and the dynamics of its evolution are intensely investigated aspects of plant reproduction. Seabuckthorn (Hippophae rhamnoides ssp. turkestanica) is an alpine shrub growing wild in certain parts of western Himalaya. The previous studies have reported heteromorphic sex chromosomes in the species and yet marker-based studies indicate high similarity between the male and female genomes. Lack of information on sexual system in the species has further complicated the situation. A systematic study was thus undertaken to understand the sexual system in seabuckthorn and to discern the extent of similarity/dissimilarity between the male and female genomes by generating a large number of markers using amplified fragment length polymorphism and representational difference analysis. Floral biology and regular monitoring of species revealed the presence of polygamomonoecious (PGM) plants in most populations at a low percentage (~2–4%). PGM plants showed low pollen production and overall low fertility, suggesting a monoecy–paradioecy pathway at function. The results of the marker study demonstrated that there are limited differences between male and female genomes and these differences were not uniform across the populations in the Leh-Ladakh region, especially when the geographical distance increases. Results also suggest that a dynamic partitioning of genomes is operational between the two genders of seabuckthorn and differences are not homogenized across the populations. Both reproductive biology-based and DNA marker-based studies indicate that genders have separated recently. The present study proposes seabuckthorn as a promising model system to study evolution of dioecy and sex determination.
Similar content being viewed by others
Introduction
Dioecy is widespread among the flowering plants. It is believed to have evolved independently from cosexuality on several occasions (Renner 2014). The evolutionary routes toward dioecy may include transitory stages, represented by some plants which bear bisexual flowers either along with staminate (andromonoecious), pistillate (gynomonoecious), or both the types (polygamomonoecious (PGM)), and exist along with fully evolved unisexual plants. Plant populations with such a gender configuration are termed subdioecious (Geber et al. 1999). Subdioecy is often maintained in a population until complete dioecy is established (Ross 1982; Barrett 2002), with a possibility of reversion to cosexuality (Charlesworth and Charlesworth 1979). Breakdown of established dioecy may also result in subdioecious condition (Ehlers and Bataillon 2007). Species harboring subdioecious condition can help in understanding the prevailing mechanism of gender determination and provide the crucial empirical evidences in tracing the evolutionary forces en route to dioecy (Maurice and Flemming 1995; Renner 2014).
Genetic and molecular studies on dioecious model systems have shown significant variation in the inheritance pattern of sex, ranging from single locus to multiple loci and sex-specific chromosomes (Charlesworth 2015). So far, 39 plant taxa have been reported to show sex chromosomes among the angiosperms (Ming et al. 2011). The evolution of heteromorphic sex chromosome is proposed to initiate with suppression of recombination at the sex determination loci, which spreads to the adjacent regions of the evolving chromosome(s). Suppression of recombination is often combined with genetic hitchhiking and other Hill Robertson effects, thereby causing accumulation of repeated sequences, enlargement of Y chromosome, and eventually degeneration of Y chromosome (Ming et al. 2011). Sex chromosomes in different species have been proposed to represent these stages beginning with two loci (Fragaria) to completely degenerated Y chromosomes (Rumex) (Charlesworth 2013; Muyle et al 2017).
Seabuckthorn has been mostly described as a dioecious species in the literature (Mabberley 2008). The species thrives well in cold deserts and in temperate and sub-tropical parts of the World including Europe, Russia, Central Asia, India, and China, (Bartish et al. 2002). In India, seabuckthorn occurs in the Himalayan region. Owing to its potential as a future crop (Ruan et al. 2004), several attempts have been made to develop reliable gender-linked SCAR (sequence characterized amplified region) markers, but these met with little success. First of such studies (Persson and Nybom 1998) reported a male-specific DNA marker, which worked only for the progeny obtained from one of the two different crosses involving a single male parent. Subsequent studies reported gender-specific markers (Sun et al. 2006; Sharma et al. 2010; Korekar et al. 2012; Jadav and Sharma 2014), but these markers were not tested on geographically isolated populations. Notably, when tested on different populations, they could not identify gender (Das et al. 2017), indicating lack of genetic uniformity in various natural populations. Misidentification of the gender and lack of information on sexual system of the species could also be another important factor adding to the prevailing confusion in this regard. A systematic study thus became a prerequisite to establish the sexual system in seabuckthorn, and to understand the extent of similarity/dissimilarity between the genomes of the two genders. The present study was carried out to understand the sexual system in natural populations of Hippophae rhamnoides growing in western Himalayan region of India. The differences between male and female genomes were investigated using molecular markers and efforts were made to understand the nature of the identified loci showing association with a particular gender. We report the prevalence of subdioecy in natural populations of H. rhamnoides in the region. We show that the genomes of two sexes are possibly more similar than expected and highlight a lack of genetic uniformity across the populations investigated in Leh-Ladakh region.
Materials and methods
Study site and material
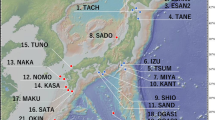
The study covered 25 natural populations of Hippophae rhamnoides ssp. turkestanica spread over three valleys namely Indus, Nubra, and Suru from Leh-Ladakh (3000–5000 m above sea level), Jammu and Kashmir, India (Supplementary data Table S1). The region experiences a mean annual temperature in the range between −12 °C and 30 °C and high wind speed. The populations were mapped by using ArcMAP 9.1 software (2007) (Fig. 1). The plants were assigned sex (male and female) based on field observations (flowers and fruits) as described in our previous studies (Mangla et al. 2013, 2015; Mangla and Tandon 2014). Some plants with sparse fruit set (as compared to female plants) were also observed. These plants were sampled separately and were later identified as PGM plants.
a Outline map of India (not to scale) showing geographical region where study was conducted. b Distribution of 25 populations across three valleys, from where the plants were sampled (see Supplementary data Table S1 for details)
Sexual system
Sexual system among the plants was established during 2009–2012, by ascertaining the functionality of flowers (floral biology), resources allocated to stamens and pistil in the various floral morphs, and crossability among them. For this study, two populations viz. Choglamsar (CV) and Water Park (WP) from Indus valley were chosen and five plants were marked in each population (Supplementary data Table S1). These populations were chosen as they were away from human establishments and free from anthropogenic activities. The plants in these populations were also monitored for the consistency in gender expression during the study period.
Floral biology
Temporal details of anthesis (opening of flowers) were recorded by monitoring the marked plants (n = 5 plants each gender, n = 50 inflorescence per plant, two populations) over a period of 15 flowering days (between April and May) for two seasons. The number of flowers borne per inflorescence (n = 75 racemes, 10 plants) was counted under a stereozoom microscope. For anatomical details and scanning electron microscopy, flowers were processed as described earlier (Mangla et al. 2013). Pollen viability and fertility were estimated by fluorochromatic reaction and acetocarmine staining (Shivanna and Tandon 2014), respectively.
Reproductive allocation
The relative proportion of resources allocated among the floral organs gives an idea about the trade-off between male and female functions (Charnov 1982). The male, female, and PGM plants (n = 10 each) were compared for the allocation, measured in terms of mean production of flowers (per inflorescence), pollen (per flower), pollen viability, pollen fertility, and fruits (per infructescence). The datasets for each parameter were pooled from each population and mean values were compared for difference using one-way analysis of variance (ANOVA) and post hoc Tukey’s test. The total male reproductive output (i.e., pollen production, pollen viability, and pollen fertility) of male flowers of PGM plants was also compared with stamens borne by the bisexual flowers of PGM plants, by using one-way ANOVA and grouped by applying post hoc Dunnett test for unequal datasets. The percentage datasets of pollen fertility and viability were normalized by square-root arc-sine transformation before analysis. The statistical analyses were carried out using the IBM SPSS statistics 21 (IBM Corp. 2012).
Breeding system: cross-compatibility among the genders
Crossability of the two genders with various floral morphs borne on the PGM plants was ascertained through controlled pollination experiments in three combinations: (i) PGM plants x male plants (n = 42), (ii) female plants x PGM plants (n = 70), and (iii) female flowers from the PGM plants x pollen grains from male flowers of PGM plants (n = 35 female flower); in order to study pollen germination and in vivo pollen tube growth, the pistils were fixed (75 h after pollination) in lactic acid:ethanol solution (1:3, v/v) and stained with decolorized aniline blue (Martin 1959) and observed under an epifluorescence microscope (Eclipse 80i Nikon, Japan).
Isolation of gender-specific markers
In the present study, two contrasting methodologies, viz. representational difference analysis (RDA) and amplified fragment length polymorphism (AFLP), generated markers in large number, which increased the chances of identifying a gender-specific marker, and also provided a measure for the extant of similarity/dissimilarity between the genomes of male and female genders. RDA generated markers based on differences between the genomes of two genders, while AFLP was carried out for samples from different populations to capture gender-specific genomic differences, which are uniform across populations.
Sampling of material and DNA isolation
A total of 175 plants (70 males, 73 females, 32 PGM) (Supplementary data Table S1) were sampled from 25 natural populations. The minimum distance between any two sampled populations was ~4 km. As seabuckthorn also reproduces asexually through root suckers, samples collected within a population were at least 15 m apart. Leaves were wrapped in aluminum foil, brought to the laboratory in liquid nitrogen, and stored at −80 °C until DNA extraction. DNA was extracted as described by Raina et al. (2012).
Representational difference analysis
Male and female DNA samples were taken from RP population; male was used as a tester and female as a driver. 2.5 μg of DNA was sheared from each tester and driver by a sonicator (Covaris S2, Massachusetts, USA) to a range of 100–500 bp. Sheared DNA was purified using gel elution kit (Qiagen, Hilden, Germany), end-repaired using end-repair kit (New England Biolabs, India), and purified again using PCR purification kit (Qiagen, Hilden, Germany). Subsequently, blunt-end MluI and StuI + NheI adapters (synthesized by IDT, USA) were ligated to the tester and driver DNA, respectively, using T4 DNA ligase enzyme (Roche, USA). Subsequent steps (PCR, denaturation, hybridization, subtraction) were done as per Liao and Tsai (1999) with three rounds of hybridization and subtraction increasing the amount of driver DNA in each subsequent round (tester:driver ratio of 1:80, 1:400, and 1:1400). To compensate the loss of tester sequences, each subtraction round was followed by amplification round of three cycles with tester-specific primer. The subtracted products were cloned and sequenced as per Bali et al. (2015).
RDA resulted in 600 positive clones, which on sequencing generated 403 good quality sequences. One hundred and eighty sequences assembled into 61 contigs (National Center for Biotechnological Information (NCBI) GenBank accessions: KS372811–KS372871), while 223 sequences remained singletons (NCBI GenBank accessions: KS372588–KS372810) ranging from 90 to 595 bp in size. The sequences were subjected to nucleotide BLAST (threshold e value: 10−8) categorizing them into four groups: (1) plastid, mitochondrion, or ribosome specific (19 contigs, 51 singlets); (2) microsatellite or various repeat sequence specific (6 contigs, 22 singlets); (3) sequences with no significant hit (25 contigs, 119 singlets); (4) sequences of genes known to play a role in sexual reproduction (11 contigs, 31 singlets). A total of 76 primer pairs were designed from all the categories, considering various aspects like the quality of reads, e value, percentage match, and the sequence length. Six primer pairs were designed from the first category, 14 from the second, 35 from the third, and 21 from the fourth one.
Amplified fragment length polymorphism
AFLP was performed as per Bali et al. (2015). The pre-amplified products were diluted (1/50) and used for selective amplification. The profiles were generated on polyacrylamide gel electrophoresis (PAGE) using LI-COR 4300 DNA analyzer (LI-COR Biosciences, Lincoln, USA) and images were viewed with Gel Buddy software. A total of 108 combinations of selective primers (EcoRI + MseI) were screened using a male and a female plant. Seventy-one primer combinations (Supplementary data Table S2) were selected and tested on 23 male and 23 female plants representing five populations (CV, RP, WP, FRL, and S).
Elution, cloning, and sequencing of fragments
The selected AFLP profiles were repeated on 6.5% PAGE using Sequi-GenR GT Nucleic acid electrophoresis cell (Bio-Rad, USA). The desired fragments were eluted as per Goel et al. (2006), and were cloned and sequenced as described earlier. A minimum of three clones were sequenced for each AFLP fragment.
Development of gender-specific SCAR markers and their validation
Based on the sequences of AFLP fragments, primers were designed and tested for their gender specificity on different natural populations. The 25 μl of PCR reaction mix contained: 100 ng of genomic DNA, 0.4 µM each of forward and reverse primer, 0.02 mM of each dNTPs, 1× PCR buffer with 2 mM MgCl2 (final concentration), 1 U of Taq DNA polymerase (Intron Biotechnology, India), and appropriate amount of water (Sigma, Missouri, USA). The PCR cycling consisted of an initial denaturation at 95 °C for 5 min followed by 30 cycles of denaturation (94 °C, 30 s), annealing (appropriate temperature, 30 s), extension (72 °C, 1 min), and a final extension (72 °C, 7 min). The amplification of rDNA Internal Transcribed Spacer (ITS 1 and ITS 4) was used as a positive control. The PCR reaction was set as per White et al. (1990). SCAR loci were cloned and sequenced as described earlier. A minimum of three clones were sequenced per amplified fragment.
Characterization of gender-specific locus
We looked around the SCAR region to understand the nature of locus and to investigate how far the locus holds gender specificity.
Genome walking
The genomic region around SCAR was explored through genome walking (both 5′ and 3′) in the plant from which the SCAR was originally isolated. Total genomic DNA (1 µg) was digested with rare DNA restriction enzymes, that is, DraI, SspI, StuI, and EcoRV (NEB, India). The adapter’s (AP1 and AP2) ligation, primary PCR conditions, and secondary PCR conditions were followed as detailed in the Genome Walker™ Universal Kit (Clontech, California, USA). Primary PCR was done using the AP1-specific primers provided in the kit and the primer designed based on sequence generated from the locus.
Gender specificity test of genome-walked region
To test gender specificity, primers were designed from different segments of the region walked (Supplementary data Table S3). These primers were tested in 50 males, 50 females, and 30 PGM samples collected across the 25 populations. The PGM plants were only tested for the presence of gender-specific marker to find possible inclination of them towards any specific gender (if any). The PCR conditions for amplification and protocols of cloning and sequencing were same as described earlier.
DNA dot blot analysis
The copy number of genome-walked fragments was assessed by dot blot hybridization. For each clone, 100 ng of plasmid DNA was heat denatured (95 °C for 5 min) and spotted on the N+ nylon membrane (Amersham, New Jersey, USA). Four hundred nanograms of autoclaved total genomic DNA from male or female plant was labeled and used as probe. Probe labeling was done with digoxigenin (DIG)-dUTP using the random prime labeling method (Roche, USA). Hybridization and detection were performed using DIG high prime labeling and detection starter kit II (Roche, USA). The images were captured with Nightowl chemiluminescence detection system (Berthold, Tennessee, USA).
Sequence analysis
The sequences were assembled using DNA Lasergene Core suite software (version 12). The sequence alignments were done using the software Clustal X (version 2). BLASTN tool was used to perform various homology searches against the non-redundant and conserved domain databases at the NCBI and Repbase database of the Genetic Information Research Institute. The putative amino acid sequences were predicted using an online translate tool, ExPASy (https://www.expasy.org/).
Results
Sexual system
Floral biology of the species and regular monitoring of the tagged plants over 4 years at the sites revealed the occurrence of three genders in seabuckthorn viz. male, female, and PGM. Out of the 25 natural populations studied, PGM plants were present in 12 (Supplementary data Table S1). The plants of three genders were consistent in their gender expression during the period of study (2009–2012) and do not exhibit sex lability.
Flowers of PGM plants
Inflorescence of seabuckthorn is a condensed axillary raceme. In PGM plants, a raceme bears 7.75 ± 1.58 flowers, which was lower than those borne by the male plants (9.29 ± 1.58) and greater than the females (6.14 ± 1.81). The pattern of anthesis within a raceme of PGM plants (0900–1200 h) was random, while in the male and female plants, it occurred (0800–1200 h) in an acropetal manner (Fig. 2a–c). The ratio of the three floral morphs viz. staminate, pistillate, and hermaphrodite in a PGM plant was 1:1:0.11. The structural organization of staminate and pistillate flowers of PGM plants were similar to those borne by the male and female plants (see Mangla et al. 2013; Mangla and Tandon 2014). Among the PGM plants, staminate flowers were invariably superior to the hermaphrodite flowers in terms of pollen production and pollen fertility (Table 1). However, ~20% of pistillate flowers were sterile and produced carpelloid-like structure, gynoecium without ovule, or a rudimentary ovary with deformed stigma (Supplementary data Fig. S1).
Inflorescence of female (a), male (b), and polygamomonoecious plants (c) showing flowers (arrow). d–f Longitudinal section of bisexual flowers (A1G, A2G, and A4G) of PGM plants. Note that conduplication is incomplete in A2G (e) and A4G (f) flowers (arrow). an: anther, sg: stigma. Scale bars: a = 2 mm, b = 6 mm, c = 5 mm, and d–f = 500 μm
The three morphs of hermaphrodite flowers were observed/characterized based on number of anthers, that is, having one, two, or four anthers along with gynoecium, henceforth termed A1G, A2G, and A4G, respectively (Fig. 2d–f). In A1G type, gynoecium is sessile and distinguishable into a dry type stigma with smooth to slightly undulated surface, a conspicuous pseudostyle, and a dorsally protruded ovary (Fig. 2d). However in A2G type, pseudostyle is absent and the intervening region between the stigma and ovary is marked by incomplete conduplication (longitudinal carpel closure; Bailey and Swamy 1951), where the carpel margins are placed wide apart. In the ovarian region, the carpel wall is loosely appressed to enclose the ovule (Fig. 2e). In A4G type, the ovarian region was completely open along the ventral surface due to partial involution of the carpel wall (Fig. 2f). In all these bisexual flowers, the stigma was mostly positioned away from the anthers. In comparison to the female flowers (our previous study, Mangla et al. 2013) there was appreciable amount (~60%) of gynoecial sterility in these three floral morphs.
The PGM plants were self-compatible. Pollen from the male plants could also germinate on the stigma of the female flowers of PGM plants. However, pollen sourced from PGM plants failed to germinate on the stigma in female plants (Supplementary data Fig. S2).
Reproductive allocation in male, female, and PGM plants
Reproductive allocation among the three genders differed significantly (Table 2). The allocation was greater among the male plants for flower production (per raceme), pollen production, pollen fertility, and viability than that in PGM plants. The male plants produced 1.5 times more flowers per raceme in comparison to female plants. The female plants were superior by producing two-fold more fruits than the PGM plants (per infructescence) (Table 2).
Isolation of gender-specific marker
Representational difference analysis
Seventy-six primer pairs were designed based on RDA as explained under Materials and methods. Out of these, only 17 could differentiate male from female plants which were used for RDA. However, only four primer pairs showed gender specificity (RDAP-27, RDAP-33, RDAP-41, and RDAP-47) when further tested on multiple samples from RP population (Indus valley). All the four sequences showed similarity to satellite or transposons when subjected to homology search (Supplementary data Table S4). RDAP-27 and RDAP-47 amplified a female-specific amplicon of ~300 bp and ~1.2 kb, respectively, while RDAP-41 generated a male-specific amplicon of size ~350 bp (Supplementary data Fig. S3). These primer pairs failed to discriminate gender when tested on additional populations, hence were excluded from further analysis.
Primer pair RDAP-33 exhibited size variation by amplifying a ~1.5 and ~1 kb amplicon in male and female samples, respectively, which were eluted and sequenced (NCBI GenBank accessions: KX444155–KX444156). The alignment of the two sequences showed high homology between them but male sequence had additional scattered insertions of 1–31 bp. Based on this alignment, a primer pair (SCAR-33) was designed which amplified a male-specific amplicon of ~1.5 kb in different populations from Indus valley (RP, UP, RP, S, CV, and FRL) and Suru valley (BA, SP, UT, and NI), although it did not hold gender specificity for the nine populations tested from Nubra valley (Supplementary data Fig. S3).
Amplified fragment length polymorphism
Screening of 71 selective primer combinations (Supplementary data Table S2) on five populations identified various gender-specific fragments. Since gender specificity of fragments in specific population could have been due to intrinsic variation available in the genomes, only those fragments were considered which could differentiate genders in all five populations. Only six primer combinations generated gender-specific fragments in all the five populations tested. Four of these fragments were female specific, while two were specific to males (Supplementary data Fig. S4). These fragments were cloned and sequenced for further analysis (Supplementary data Table S5).
Fragment sequencing, development of SCAR marker, and its validation
Cloning and sequencing of the above-mentioned six fragments revealed 10 different sequences. Two fragments with primer combinations E-AGG + M-CAGT and E-ACC + M-CTG revealed three different sequences each. Rest of the four fragments generated only one type of sequence each (NCBI GenBank accessions: KX444157–KX444166). The nucleotide BLAST results are shown in Supplementary data Table S5. Based on these sequences, a total of 10 primers pairs were designed and initially tested on male, female, and PGM plants representing six different populations of Indus valley (CV, RP, SD, WP, S, FRL). Seven primer pairs did not show gender specificity and were not investigated further. Three primers pairs (H. rhamnoides male locus (HRML), AFLP-32, and AFLP-15) were further validated on males, females, and PGM plants from additional 19 populations from three geographically isolated valleys.
Primer pair SCAR-32 (from AFLP-32) (NCBI GenBank accession: KX444164) produced amplicons of different sizes in the male (225 bp) and female (213 bp) plants of RP and S (Indus valley) population but did not hold the gender specificity when tested on other 17 populations (Indus valley: CV, WP, FRL, NI, UP; Nubra valley: TK, SR, KS, KR, SU1, SU2, SU3 HS, DK; Suru valley: BA, SP, UT) (Supplementary data Fig. S5-A). The alignment of amplicon sequence from four populations (RP, SU1, SU2, UT) showed fair amount of conservation with minor differences, suggesting that similar locus has amplified across the valleys. A 12 bp insertion in females was responsible for the observed size difference (Supplementary data Fig. S5-B).
The fragment generated from E-AGG + M-CTG (AFLP-15) was female specific and only 67 bp in size, which was not sufficient to design SCAR primers. Genome walking was performed in both 5′ and 3′ direction adding 321 and 158 bp to this fragment (NCBI GenBank accession: KX444165) enabling designing of primer pairs (Supplementary data Table S5). When the amplification was tested with the primers (SCAR-15) on various populations, varying results were obtained. In some populations, an ~250 bp fragment amplified either in male (Indus valley: NI; Nubra valley: SU3, DK; Suru valley: SP, UT) or female samples (RP), while in other populations (Indus valley: S, TK, Nubra valley: SR, KS, KR, SU1, SU2), an ~250 bp fragment was amplified in both male and female plants. No amplification was obtained in BA (Suru valley), UP2 and UP3 (Indus valley) populations.
Among all the three AFLP markers, only HRML produced a male-specific 329 bp fragment (Supplementary data Table S5) across the 25 populations of three valleys (Supplementary Fig. S6-A). This SCAR also amplified across 60% of PGM plants tested. The amplified fragment from the male plant of 11 different populations (Indus valley: CV, SD, WP, FRL; RP, NI, UP3; Nubra valley: HS, SU2, KS, SR) was eluted, cloned, and sequenced. No sequence divergence was observed at the locus across the populations (Supplementary data Fig. S5-B; NCBI GenBank accessions: KX444166–KX444176).
Characterization of HRML locus and flanking regions
Success of HRML as a gender-specific SCAR marker prompted us to further characterize this locus through genome walking, thereby increasing the length of locus by ~3.8 kb in 5′ direction and ~3.1 kb in 3′ direction, and resulting in an ~7 kb region named as HRMSSR (H. rhamnoides sex-specific region, NCBI GenBank accession: KX444194).
The sequence of HRMSSR was compared across the populations by designing different primers (Supplementary data Table S3). Most of the primers generated fragments of same size in all male and female samples barring two primer pairs (Fig. 3a). First primer pair (HRML1 and HRML3) amplified a fragment of ~676 bp in all the male samples and in many (90%) PGM plants, but amplification was absent in the female plants (Supplementary data Fig. S7-A). The amplicons were eluted from five male plants (one each from five population viz. Indus valley: CV, WP, SD; Nubra valley: SU1, SU2), cloned, and sequenced. The sequence did not show any divergence at this locus (Supplementary data Fig. S7-B; NCBI accession no: KX444177–KX444181).
Diagrammatic representation of HRMSSR locus (1–7016bp). a Gel profiles showing amplifications in male (left lane) and female (right lane) samples tested with different primer pairs (Supplementary data Table S3). Black bars show the male-specific amplification across populations with primer pairs HRML1 and HRML2 and HRML1 and HRML3. Lined gray block represents the amplicon of ~1200 bp in female with primer pair HRSS1 and HRML2 (see results for detail). b Location of a satellite, microsatellite, and transposons in HRMSSR locus. Dotted arrows show the position of 5 bp direct repeats “ACTCT” in female samples. Male has only one 5 bp sequence (ACTCT). c Organization of Ty3-gypsy-like and LTR-Ty1-copia-like retroelements at HRMSSR locus. The snapshots of alignments show conserved motifs (*). Black solid circles denotes the stop codons in translated HRMSSR sequence. Earlier known transposons sequences used for the alignment were obtained from the NCBI GenBank database. {Tobacco (Tto1): BAA11674, D83003; Tobacco (Tnt1): CAA32025, X13777; Orange (CIRE1): CAJ09951, AM040263; Drosophila (Copia): CAA26444, X02599; Tomato (TLC1.1): AAK29467, AF279585; Malus domestica (Ctrm1): FJ705357, XP00837822; Noccaea caerulescens (TNT1-94): JAU86839.1; Cajanus cajan: KYP58323.1; Phoenix dactylifera: XP_008779305.1; Populaus euphratica: XP_011027642.1; Ziziphus jujuba: XP_015894632.1; Hordeum vulgare: AY040832.1; Solanum lycopersicum: AAD13304.1; Arabidopsis thaliana: AAC69377.1; Zea mays: AAL76001.1}
The second primer pair (HRSS1 and HRML2) produced amplicons of differing size between male (1100 bp) and female plants (1200 bp) (Supplementary data Fig. S8-A). PGM plants produced a fragment of either ~1200 or ~1100 bp. The alignment of consensus nucleotide sequences of amplified products from one male and one female plant each from six different populations (Indus valley: SD, CV; Nubra valley: HS, DK, KS, SU1) showed an insertion of about 95 bp in the female plants. Interestingly, this insertion was splitting the SCAR primer HRML1 (primer designed based on an AFLP sequence) in females (NCBI GenBank accessions: KX444182–KX444193) resulting in a male-specific amplification (Supplementary data Fig. S8-B).
Homology of HRMSSR sequence
Homology search of HRMSSR contig revealed that HRML locus is flanked by two inactive retrotransposons, that is, Ty3-gypsy at the 5′ end and Ty1-copia at the 3′ end (Fig. 3b). Transposon’s inactive status was evident by multiple stop codons, frameshift mutations, incomplete long terminal repeat (LTR), and so on. Ty3-gypsy element extended from 1 to ~3100 bp and resembled the evolutionary lineages, Chromovirus. It included conserved motifs for enzymes reverse transcriptase (RT), RNase H (RNASE), integrase (INT), and chromodomain (CD) in order of 5′-RT-RNASE-INT-CD-3′, characteristic of a typical Ty3-gypsy chromovirus lineage. The RT enzyme region showed conservation of four of the five amino acid residues SKCEF and seven of eight residues of putative nucleic acid-binding domain, GLRSGY-F (Fig. 3c). The conserved signature motif of RNase enzyme, that is, T/CDAS was not visible, although the three distinct conserved motifs of INT enzyme were found intact. Adjacent to the INT domain, a mutated CD region was also evident. The region following the CD was expected to contain 3′ LTR of the Ty3-gypsy retrotransposon. However, we were not able to trace the boundaries of the putative 3′ LTR owing to the loss of structural features and the absence of 5′ LTR of Ty3-gypsy retrotransposon. A modified zinc-finger domain H10H29C2C at the N terminal, the central GLLQPLPI motif, and the characteristic catalytic motif D60D35E were also identified. In the adjoining region of the Ty3-gypsy retrotransposon, a mini satellite repeat (TTTACTCATTGAATTAAGGATTCCTTGT)7 and a perfect microsatellite repeat (ATT)20 were noted at the nucleotide position 2153 and 2605 bp, respectively (Fig. 3b).
Ty1-copia retrotransposon extended from 4499 to 6713 bp. Analysis of Ty1-copia retrotransposon identified a putative 12 bp tRNAmet primer binding site (PBS) at 4970 bp which is required for first-strand synthesis during reverse transcription of the retrotransposable elements (Fig. 3c). The PBS sequence was similar to that reported in other plant species. The characteristic 3′ inverted “CA” repeat motif of the putative 5′ LTR of Ty1-copia retrotransposon was present prior to the PBS. The putative 5′ LTR consisted of several sequence features including TATA box (TATATATTA), CAAT box, polyadenylation, and termination stem loop signals. However, the 5′ end of the LTR could not be delineated due to the truncated nature of isolated element. Downstream to the PBS region, gag domain followed by gag-pre INT domain of Ty1-copia element were recognized. A conserved nucleic acid-binding motif, C2C4H4C, was also observed in the gag region of partial Ty1-copia retrotransposon. Further downstream, a partial and modified zinc-finger motif of Ty1-copia INT enzyme (H4H19C2C) was also present.
DNA dot blot analysis
Clones representing different sequenced regions of HRMSSR were dot bloted and hybridized seperately with male or female genomic DNA (Supplementary Fig.S9). The sequences from region 947–2693, 2694–3475, 3953–5002, and 5864–7016 showed moderately high copy number with both male and female genomes.
Discussion
The present study attempts to understand evolution of dioecy in seabuckthorn by analyzing natural populations distributed over three valleys in Leh-Ladakh region. The study established the subdioecious sexual system in the species at the sites through field observations, and also compared the genomes of male and female individuals using two versatile DNA markers viz. RDA and AFLP.
Subdioecy in seabuckthorn
Contrary to earlier reports (e.g., Singh 2003; Korekar et al. 2012, Raina et al. 2012), the natural populations of seabuckthorn from western Himalaya investigated in the present study were found to be subdioecious. The populations were comprised of a mixture of strictly unisexual and PGM plants; the proportion of latter ranged from ~2 to 4%.
PGM plants in general are expected to have high fruit set through enhanced opportunities of self-fertilization. However, in seabuckthorn the PGM plants exhibited limited fruit set due to low pollen production, low fertility (both male and female) (see Table 2), indicating that self-pollination may not be a favored strategy in seabuckthorn. The inability of the PGM plants to equally invest in two genders highlights that (1) there is trade-off between male and female functions, and (2) they are reproductively inferior, hence maintain their genotypes in the population at low frequency (Charlesworth and Charlesworth 1978a, 1981). Additionally, the nonfunctional, sterile, and rudimentary gynoecium in some female and hermaphrodite flowers and partial pollen fertility observed in the PGM plants indicate a comparatively recent and ongoing divergence of sexes (dioecy) in seabuckthorn. This is also supported by high similarity between the male and female genomes as discussed later.
Dioecy can evolve from a hermaphrodite ancestor primarily through two pathways, that is, gynodioecy and monoecy (Barrett 2002), which can be distinguished based on the intermediate stages of sexuality at the population level. In the gynodioecious pathway, the transitory stage involves populations exhibiting variability in sexual function of females and hermaphrodites, while in the monoecy–paradioecy pathway, the transitional population exhibit bimodality of gender and possesses quantitative variation in female and male fertility (Charlesworth and Charlesworth 1978b; Spigler and Ashman 2012). Several findings of the present work support the prevalence of hermaphrodite–monoecy–dioecy pathway in seabuckthorn. First, PGM plants in seabuckthorn produce significantly lower amount of pollen grains with low viability and fertility than the males (see Table 2). The establishment of male forms of PGM plants with low pollen output, high male sterility, and reduced self-fertilization in bisexuals among the seabuckthorn populations indicates the presence of this pathway. Second, in controlled pollination experiment involving PGM plants and unisexual plants of seabuckthorn, PGM pollen failed to germinate on the stigma of female plants. This cross-incompatibility between unisexual females and PGM plants limit the male function of PGM plants and fulfill an essential requirement for the establishment of dioecy through monoecy. Had there been PGM pollen germination on the females, it would have supported the existence of a “gynodioecious pathway”. Third, bisexual flowers of PGM plants showed variable number of anthers (i.e., A1G, A2G, and A4G type) and gynoecium with structural variability. Such a stage with range of co-sexual variants suggests a gradual reduction in the male and female functions during the early stages of evolution of dioecy through monoecy (Charlesworth and Charlesworth 1978a, 1978b, 1979). This could be a manifestation of relative proportions of maleness and femaleness in bisexual flowers due to partial sterility mutations expected in hermaphrodite–monoecy–dioecy pathway (Geber et al. 1999).
Limited differences between male and female genomes
The present study generated a large number of markers highlighting the lack of differences between male and female genomes, as not many gender-specific markers were identified. This high similarity observed between male and female genomes suggests a recent/ongoing evolution of dioecy in seabuckthorn. Earlier studies, based on their attempts to isolate gender-specific SCAR markers, have also suggested high homology between the genomes of male and female plants in H. rhamnoides (Persson and Nybom 1998; Sheng et al. 2006; Sun et al. 2006; Sharma et al. 2010; Korekar et al. 2012; Das et al. 2017). These studies were based on limited efforts using markers like RAPD, SSR etc., and emphasized the need to generate a large number of markers to understand the extent of similarity between male and female genomes. The two marker systems used in the present study generated large number of markers, yet fewer than the expected gender-specific markers were identified (Supplementary Table S3), reiterating that there are limited genomic differences between male and female genomes.
Differences between male and female genomes are not uniform across populations
Besides the lack of differences between male and female genomes, present study also emphasized that the limited differences observed between male and female genomes were also not uniform across the populations studied. The differences identified between the male and female genomes can be categorized as “within population” and “between populations.” Within a population, gender-specific markers were present indicating that dynamic partitioning of genomes between the two genders is operational, but these markers lose their gender specificity at inter-population level. RDA was able to capture differences between the two genomes and identified a large number of markers, which were population specific; however, only one (SCAR-33) marker could distinguish the genders among plants from different populations of two valleys viz. Indus and Suru valley, but could not differentiate gender in populations from Nubra valley (Supplementary data Fig. S3-C, D). Similarly in AFLP, among the 71 selective primer pairs tested, only 6 could generate fragments distinguishing the gender across populations. Among these only one marker (HRML) could distinguish the genders in populations tested from all the three valleys. Such population-specific behavior of gender-specific loci could be due to limited pollen flow and reproductive isolation through geographical barriers (Mangla and Tandon 2014; Raina et al. 2012), due to which distantly located populations are less likely to exchange alleles. This fact is more pronounced with the increased physical distance and is at its maximum when populations between different valleys are considered. In contrast, alignment result of HRML locus from the different populations showed high conservation of the sequences across the valleys (Supplementary data Fig. S6B). It is likely that this locus had become fixed well before the spread of species over this large geographical area and might be playing an important role in sex determination. Considering this we characterized this locus in more details.
Characterization of gender-linked SCAR locus
HRML locus extended through genome walking and was termed as HRMSSR. HRMSSR region was compared between male and female individuals from the three valleys (Fig. 3a). Approximately 1500 bp of HRMSSR locus was compared though sequencing and differed only by additional ~95 bp in case of females (Fig. 3b). The other regions of HRMSSR were compared between the two genders through PCR and did not show any difference in their amplification pattern (Fig. 3a). Earlier studies have shown presence of heteromorphic X and Y chromosomes in seabuckthorn (Shchapov 1979; Elena et al. 2011; Puterova et al. 2017), suggesting that there should be a significant level of difference between male and female genomes (Hyvönen 1996). However, the present study does not support this notion. Similarity between the two genders at gender-specific loci can also increase the chances of occasional recombination (Ming et al. 2011), which may lead to breakdown of dioecy and manifestation of subdioecy in seabuckthorn. A similar condition has also been reported in case of Asparagus officinalis and Spinacia oleracea, where hermaphroditic flowers or monoecious plants are occasionally produced due to recombination between sex-determining regions (Khattak et al. 2006; Telgmann-Rauber et al. 2007). Additional studies are required in seabuckthorn to investigate the location of gender-specific loci identified in the present work, to ascertain their presence on autosomes/sex chromosomes.
Abundance of repeats and transposons at gender-specific loci
All the gender-linked markers generated in the present study showed homology with repeats and retrotransposons. Detailed analysis of HRMSSR locus showed the presence of repeat sequences and retrotransposons both in male and female plants. A small deletion/insertion of ~95 bp is the only difference observed at HRMSSR locus when compared between the male and female plants. Interestingly, this ~95 bp sequence in females is flanked by a small direct repeat of 5 bp. A direct repeat can be an indication of DNA transposon activity, often generated when transposon excise from the insertion point. A direct repeat can also cause deletion of the intervening portion and could be the reason for absence of this sequence in case of male plants (Feschotte and Pritham 2007). A recent study in H. rhamnoides has also confirmed the abundance of Ty3-gypsy and Ty1-copia LTR retrotransposons in the genome of the H. rhamnoides (Puterova et al. 2017).
The present study is the first of its kind in Himalayan seabuckthorn. Our study proposes that although there is high similarity between male and female genomes, dynamic partitioning of genome is operational in the species under conditions conducive for wind pollination, a strong ecological correlate of dioecy (Renner 2014). Various population-specific gender-linked markers have been characterized, but they are neither homogenized among populations nor across the valleys investigated. These observations suggest that the genders have recently separated and are at an incipient stage of genome differentiation in H. rhamnoides ssp. turkestanica. We propose seabuckthorn to be a promising model system to study and test the hypothesis of evolution of dioecy and sex determination mechanism in a land plant occurring in extreme ecological conditions.
Data archiving
RDA-derived 61 contigs and 223 singleton sequences have been submitted to NCBI GenBank (NCBI GenBank accessions: for singletons KS372588–KS372810; for assembled contigs KS372811–KS372871). All AFLP-derived and RDA-derived assembled sequences and so on are also submitted and their NCBI GenBank accessions are KX444157–KX444194.
References
Bailey IW, Swamy BGL. (1951) The conduplicate carpel of dicotyledons and its initial trends of specialization. American Journal of Botany 38:373–379
Bali S, Mamgain A, Raina SN, Yadava SK, Bhat V, Das S et al. (2015) Construction of a genetic linkage map and mapping of drought tolerance trait in Indian beveragial tea. Mol Breed 35:112–131
Barrett SCH (2002) The evolution of plant sexual diversity. Nat Rev Genet 3:274–284
Bartish IV, Jeppson N, Nybom H, Swenson U (2002) Phylogeny of Hippophae (Elaeagnaceae) inferred from parsimony analysis of chloroplast DNA and morphology. Syst Bot 27:41–54
Charlesworth D (2013) Plant sex chromosome evolution. J Exp Bot 64:405–420
Charlesworth D (2015) Plant contributions to our understanding of sex chromosome evolution. New Phytol 208:52–65
Charlesworth B, Charlesworth D (1978a) A model for the evolution of dioecy and gynodioecy Am Nat 112:975–997
Charlesworth D, Charlesworth B (1978b) Population genetics of partial male-sterility and the evolution of monoecy and dioecy. Heredity 41:137–153
Charlesworth D, Charlesworth B (1979) The evolutionary genetics of sexual systems in flowering plants. Proc R Soc Lond Ser B 205:513–530
Charlesworth D, Charlesworth B (1981) Allocation of resources to male and female functions in hermaphrodites. Biol J Linn Soc 15:57–74
Charnov EL (1982) The theory of sex allocation. Princeton University Press, Princeton, NJ
Das K, Ganie SH, Mangla Y, Dar TU, Chaudhary M, Thakur RK et al. (2017) ISSR markers for gender identify and genetic diagnosis of Hippophae rhamnoides ssp. turkestanica growing at high altitudes in Ladakh region (Jammu and Kashmir). Protoplasma 254:1063–1077
Ehlers BK, Bataillon T (2007) “Inconstant males” and the maintenance of labile sex expression in subdioecious plants. New Phytol 174:194–211
Elena T, Capraru G, Rosu CM, Zamfirache MM, Olteanu Z, Manzu C (2011) Morphometric pattern of somatic chromosomes in three Romanian seabuckthorn genotypes. Caryologia 64:189–196
Feschotte C, Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41:331–368
Geber MA, Dawson TE, Delph LF (1999) Gender and sexual dimorphism in flowering plants. Springer, Germany
Goel S, Zhenbang C, Yukio A, Conner JA, Basu M, Gualtier G et al. (2006) Comparative physical mapping of the apospory-specific genomic region in two apomictic grasses: Pennisetum squamulatum and Cenchrus ciliaris. Genetics 173:389–400
Hyvönen J (1996) On phylogeny of Hippophae (Elaeagnaceae). Nord J Bot 16:51–62
Jadav MS, Sharma TR (2014) Identification of gender specific DNA markers in sea buckthorn (Hippophae rhamnoides L.). Indian Res J Genet Biotech 6:464–469
Khattak J, Torp A, Anderson S (2006) A genetic linkage map of Spinacia oleracea and localization of a sex determination locus. Euphytica 148:311–318
Korekar G, Sharma R, Kumar K, Meenu, Bisht NC, Srivastava RB et al. (2012) Identification and validation of sex-linked SCAR markers in dioecious Hippophae rhamnoides L. (Elaeagnaceae). Biotech Lett 34:973–978
Liao HW, Tsai HJ (1999) A highly sensitive and simple method of genomic subtraction. J Fish Soc Taiwan 26:153–159
Mabberley DJ (2008) Mabberley’s Plant-Book: a portable dictionary of plants, their classification and uses, 3rd edn. Cambridge University Press, p 1021, Cambridge, UK.
Mangla Y, Tandon R (2014) Pollination ecology of Himalayan sea buckthorn, Hippophae rhamnoides L. (Elaeagnaceae). Curr Sci 106:1731–1735
Mangla Y, Chaudhary M, Gupta H, Thakur R, Goel S, Raina SN et al. (2015) Facultative apomixis and development of fruit in a deciduous shrub with medicinal and nutritional uses. AoB Plants 7:1–12
Mangla Y, Tandon R, Goel S, Raina SN (2013) Structural organization of the gynoecium and pollen tube path in Himalayan sea buckthorn, Hippophae rhamnoides (Elaeagnaceae). AoB Plants 5:1–11
Martin FM (1959) Staining and observing pollen tubes in the style by means of fluorescence. Stain Technol 34:436
Maurice S, Flemming TH (1995) The effect of pollen limitation on plant reproductive systems and the maintenance of sexual polymorphisms. Oikos 74:55–60
Ming R, Bendahmane A, Renner SS (2011) Sex chromosomes in land plants. Annu Rev Plant Biol 62:485–514
Muyle A, Shearn R, Marais G (2017) The evolution of sex chromosomes and dosage compensation in plants. Genome Biol Evol 9:627–645
Persson HA, Nybom H (1998) Genetic sex determination and RAPD marker segregation in the dioecious species sea buckthorn (Hippophae rhamnoides L.). Hereditas 129:45–51
Puterova J, Razumova O, Martinek T, Alexandrov O, Divashuk M, Kubatet Z et al. (2017) Satellite DNA and transposable elements in seabuckthorn (Hippophae rhamnoides), a dioecious plant with small Y and large X chromosomes. Genome Biol Evol 9:197–212
Raina SN, Jain S, Sehgal D, Kumar A, Dar TH, Bhat V et al. (2012) Diversity and relationships of multipurpose seabuckthorn (Hippophae L.) germplasm from the Indian Himalayas as assessed by AFLP and SAMPL markers. Genet Res Crop Evol 59:1033–1053
Renner SS (2014) The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. Am J Bot 101:1588–1596
Ross MD (1982) Five evolutionary pathways to subdioecy. Am Nat 119:297–318
Ruan C, Qin P, Zheng J, He Z (2004) Genetic relationships among some cultivars of sea buckthorn from China, Russia and Mongolia based on RAPD analysis. Sci Hortic 101:417–426
Sharma A, Zinta G, Rana S, Shirko P (2010) Molecular identification of sex in Hippophae rhamnoides L. using isozyme and RAPD markers. For Stud China 12:62–66
Shchapov NS (1979) On the karyology of Hippophae rhamnoides L. Tsitologiya. Genetika 13:45–47
Sheng HM, An LZ, Chen T, Xu SJ, Liu GX, Zheng XL et al. (2006) Analysis of the genetic diversity and relationships among and within species of Hippophae (Elaeagnaceae) based on RAPD markers. Plant Syst Evol 260:25–37
Shivanna KR, Tandon R (2014) Reproductive ecology of flowering plants: a manual. Springer, India
Singh V (2003) Seabuckthorn (Hippophae L.)—a multipurpose wonder plant, vol 1. Indus Publishing Company, India
Spigler RB, Ashman TL (2012) Gynodioecy to dioecy: are we there yet? Ann Bot 109:531–543
Sun K, Chen W, Ma R, Chen X, Li A, Ge S (2006) Genetic variation in Hippophae rhamnoides ssp. sinensis (Elaeagnaceae) revealed by RAPD markers. Biochem Genet 44:186–197
Telgmann-Rauber A, Jamsari A, Kinney MS, Pires JC, Jung C (2007) Genetic and physical maps around the sex-determining M-locus of the dioecious plant asparagus. Mol Genet Genom 278:221–234
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA for phylogenetics. In: Innis MA, Gelfand DH, Snisky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, p 315–322
Acknowledgements
YM is thankful to the Department of Biotechnology (DBT) and Council for Scientific and Industrial Research (CSIR), Government of India, for the Junior and Senior Research Fellowships, respectively. KD acknowledges the Research Fellowships received from DBT, University Grants Commission and Ministry of Social Justice and Empowerment, Government of India. This work was financially supported by (DBT) (No. BT/PR10800/NDB/51/172/2008), Government of India and R&D grants from University of Delhi in the form of projects sanctioned to SG and RT. We are thankful to the two anonymous reviewers for their constructive suggestions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
These authors contributed equally: Yash Mangla, Kamal Das.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Mangla, Y., Das, K., Bali, S. et al. Occurrence of subdioecy and scarcity of gender-specific markers reveal an ongoing transition to dioecy in Himalayan seabuckthorn (Hippophae rhamnoides ssp. turkestanica). Heredity 122, 120–132 (2019). https://doi.org/10.1038/s41437-018-0084-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-018-0084-z