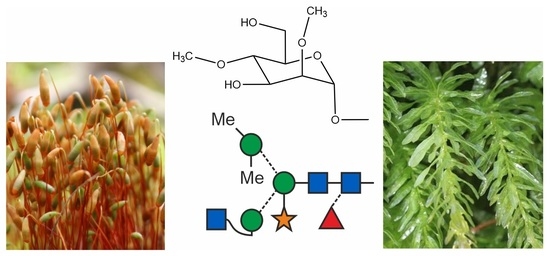
O-methylated N-glycans Distinguish Mosses from Vascular Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. N-glycan Isolation and Fractionation
2.3. Mass Spectrometric Methods
2.4. Phylogenetic Analysis
3. Results
3.1. Discovery of Methylation of Moss N-glycans
3.2. N-glycosylation Pattern of Different Moss Species
3.3. Structural Analysis of the Main Glycans from Funaria hygrometrica and Plagiomnium undulatum
3.4. Other Glycan Masses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Hykollari, A.; Paschinger, K.; Wilson, I.B.H. Negative-mode mass spectrometry in the analysis of invertebrate, fungal, and protist N-glycans. Mass Spectrom. Rev. 2021, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Schauer, R.; Kamerling, J.P. Exploration of the Sialic Acid World. Adv. Carbohydr. Chem. Biochem. 2018, 75, 1–213. [Google Scholar] [CrossRef]
- Wilson, I.B.; Zeleny, R.; Kolarich, D.; Staudacher, E.; Stroop, C.J.; Kamerling, J.P.; Altmann, F. Analysis of Asn-linked glycans from vegetable foodstuffs: Widespread occurrence of Lewis a, core alpha1,3-linked fucose and xylose substitutions. Glycobiology 2001, 11, 261–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strasser, R. Plant protein glycosylation. Glycobiology 2016, 26, 926–939. [Google Scholar] [CrossRef] [Green Version]
- Ishihara, H.; Takahashi, N.; Oguri, S.; Tejima, S. Complete structure of the carbohydrate moiety of stem bromelain. An application of the almond glycopeptidase for structural studies of glycopeptides. J. Biol. Chem. 1979, 254, 10715–10719. [Google Scholar] [CrossRef]
- Lommerse, J.P.; Kroon-Batenburg, L.M.; Kamerling, J.P.; Vliegenthart, J.F. Conformational analysis of the xylose-containing N-glycan of pineapple stem bromelain as part of the intact glycoprotein. Biochemistry 1995, 34, 8196–8206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurosaka, A.; Yano, A.; Itoh, N.; Kuroda, Y.; Nakagawa, T.; Kawasaki, T. The structure of a neural specific carbohydrate epitope of horseradish peroxidase recognized by anti-horseradish peroxidase antiserum. J. Biol. Chem. 1991, 266, 4168–4172. [Google Scholar] [CrossRef]
- Maeda, M.; Kamamoto, M.; Hino, K.; Yamamoto, S.; Kimura, M.; Okano, M.; Kimura, Y. Glycoform analysis of Japanese cedar pollen allergen, Cry j 1. Biosci. Biotechnol. Biochem. 2005, 69, 1700–1705. [Google Scholar] [CrossRef]
- Maeda, M.; Kimura, Y. Glycoform analysis of N-glycans linked to glycoproteins expressed in rice culture cells: Predominant occurrence of complex type N-glycans. Biosci. Biotechnol. Biochem. 2006, 70, 1356–1363. [Google Scholar] [CrossRef] [Green Version]
- Koprivova, A.; Altmann, F.; Gorr, G.; Kopriva, S.; Reski, R.; Decker, E.L. N-Glycosylation in the moss Physcomitrella patens is organized similarly to that in higher plants. Plant Biol. 2003, 5, 582–591. [Google Scholar] [CrossRef]
- Bencurova, M.; Hemmer, W.; Focke-Tejkl, M.; Wilson, I.B.; Altmann, F. Specificity of IgG and IgE antibodies against plant and insect glycoprotein glycans determined with artificial glycoforms of human transferrin. Glycobiology 2004, 14, 457–466. [Google Scholar] [CrossRef] [Green Version]
- Holzweber, F.; Svehla, E.; Fellner, W.; Dalik, T.; Stubler, S.; Hemmer, W.; Altmann, F. Inhibition of IgE binding to cross-reactive carbohydrate determinants enhances diagnostic selectivity. Allergy 2013, 68, 1269–1277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tretter, V.; Altmann, F.; Kubelka, V.; Marz, L.; Becker, W.M. Fucose alpha 1,3-linked to the core region of glycoprotein N-glycans creates an important epitope for IgE from honeybee venom allergic individuals. Int. Arch. Allergy Immunol. 1993, 102, 259–266. [Google Scholar] [CrossRef]
- Fitchette-Laine, A.C.; Gomord, V.; Cabanes, M.; Michalski, J.C.; Saint Macary, M.; Foucher, B.; Cavelier, B.; Hawes, C.; Lerouge, P.; Faye, L. N-glycans harboring the Lewis a epitope are expressed at the surface of plant cells. Plant J. 1997, 12, 1411–1417. [Google Scholar] [CrossRef] [PubMed]
- Melo, N.S.; Nimtz, M.; Conradt, H.S.; Fevereiro, P.S.; Costa, J. Identification of the human Lewis(a) carbohydrate motif in a secretory peroxidase from a plant cell suspension culture (Vaccinium myrtillus L.). FEBS Lett. 1997, 415, 186–191. [Google Scholar] [CrossRef] [Green Version]
- Beihammer, G.; Maresch, D.; Altmann, F.; Van Damme, E.J.M.; Strasser, R. Lewis A Glycans Are Present on Proteins Involved in Cell Wall Biosynthesis and Appear Evolutionarily Conserved Among Natural Arabidopsis thaliana Accessions. Front. Plant Sci. 2021, 12, 630891. [Google Scholar] [CrossRef]
- Koprivova, A.; Stemmer, C.; Altmann, F.; Hoffmann, A.; Kopriva, S.; Gorr, G.; Reski, R.; Decker, E.L. Targeted knockouts of Physcomitrella lacking plant-specific immunogenic N-glycans. Plant Biotechnol. J. 2004, 2, 517–523. [Google Scholar] [CrossRef]
- Strasser, R.; Altmann, F.; Mach, L.; Glossl, J.; Steinkellner, H. Generation of Arabidopsis thaliana plants with complex N-glycans lacking beta1,2-linked xylose and core alpha1,3-linked fucose. FEBS Lett. 2004, 561, 132–136. [Google Scholar] [CrossRef] [Green Version]
- Parsons, J.; Altmann, F.; Arrenberg, C.K.; Koprivova, A.; Beike, A.K.; Stemmer, C.; Gorr, G.; Reski, R.; Decker, E.L. Moss-based production of asialo-erythropoietin devoid of Lewis A and other plant-typical carbohydrate determinants. Plant Biotechnol. J. 2012, 10, 851–861. [Google Scholar] [CrossRef]
- Strasser, R.; Altmann, F.; Glossl, J.; Steinkellner, H. Unaltered complex N-glycan profiles in Nicotiana benthamiana despite drastic reduction of beta1,2-N-acetylglucosaminyltransferase I activity. Glycoconj. J. 2004, 21, 275–282. [Google Scholar] [CrossRef]
- von Schaewen, A.; Sturm, A.; O’Neill, J.; Chrispeels, M.J. Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 1993, 102, 1109–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaulfurst-Soboll, H.; Mertens-Beer, M.; Brehler, R.; Albert, M.; von Schaewen, A. Complex N-Glycans Are Important for Normal Fruit Ripening and Seed Development in Tomato. Front. Plant Sci. 2021, 12, 635962. [Google Scholar] [CrossRef] [PubMed]
- Hodgetts, N.G.; Söderström, L.; Blockeel, T.L.; Caspari, S.; Ignatov, M.S.; Konstantinova, N.A.; Lockhart, N.; Papp, B.; Schröck, C.; Sim-Sim, M.; et al. An annotated checklist of bryophytes of Europe, Macaronesia and Cyprus. J. Bryol. 2020, 42, 1–116. [Google Scholar] [CrossRef]
- Grass, J.; Pabst, M.; Kolarich, D.; Poltl, G.; Leonard, R.; Brecker, L.; Altmann, F. Discovery and structural characterization of fucosylated oligomannosidic N-glycans in mushrooms. J. Biol. Chem. 2011, 286, 5977–5984. [Google Scholar] [CrossRef] [Green Version]
- Mocsai, R.; Figl, R.; Sutzl, L.; Fluch, S.; Altmann, F. A first view on the unsuspected intragenus diversity of N-glycans in Chlorella microalgae. Plant J. 2020, 103, 184–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciucanu, I.; Kerek, F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 1984, 131, 209–217. [Google Scholar] [CrossRef]
- Cole, T.C.H.; Hilger, H.H.; Goffinet, B. Bryophyte Phylogeny Poster (BPP). PeerJ Prepr. 2019, 7, e27571v3. [Google Scholar] [CrossRef]
- Harvey, D.J. Fragmentation of negative ions from carbohydrates: Part 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass Spectrom. 2005, 16, 647–659. [Google Scholar] [CrossRef] [Green Version]
- Beike, A.K.; Lang, D.; Zimmer, A.D.; Wust, F.; Trautmann, D.; Wiedemann, G.; Beyer, P.; Decker, E.L.; Reski, R. Insights from the cold transcriptome of Physcomitrella patens: Global specialization pattern of conserved transcriptional regulators and identification of orphan genes involved in cold acclimation. New Phytol. 2015, 205, 869–881. [Google Scholar] [CrossRef] [Green Version]
- Resemann, H.C.; Herrfurth, C.; Feussner, K.; Hornung, E.; Ostendorf, A.K.; Gomann, J.; Mittag, J.; van Gessel, N.; Vries, J.; Ludwig-Muller, J.; et al. Convergence of sphingolipid desaturation across over 500 million years of plant evolution. Nat. Plants 2021, 7, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Mathieu-Rivet, E.; Mati-Baouche, N.; Walet-Balieu, M.L.; Lerouge, P.; Bardor, M. N- and O-Glycosylation Pathways in the Microalgae Polyphyletic Group. Front. Plant Sci. 2020, 11, 609993. [Google Scholar] [CrossRef] [PubMed]
- Staudacher, E. Mollusc N-glycosylation: Structures, Functions and Perspectives. Biomolecules 2021, 11, 1820. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stenitzer, D.; Mócsai, R.; Zechmeister, H.; Reski, R.; Decker, E.L.; Altmann, F. O-methylated N-glycans Distinguish Mosses from Vascular Plants. Biomolecules 2022, 12, 136. https://doi.org/10.3390/biom12010136
Stenitzer D, Mócsai R, Zechmeister H, Reski R, Decker EL, Altmann F. O-methylated N-glycans Distinguish Mosses from Vascular Plants. Biomolecules. 2022; 12(1):136. https://doi.org/10.3390/biom12010136
Chicago/Turabian StyleStenitzer, David, Réka Mócsai, Harald Zechmeister, Ralf Reski, Eva L. Decker, and Friedrich Altmann. 2022. "O-methylated N-glycans Distinguish Mosses from Vascular Plants" Biomolecules 12, no. 1: 136. https://doi.org/10.3390/biom12010136