- 1Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia
- 2State Environmental Protection Budgetary Institution of Moscow “Mospriroda”, Moscow, Russia
- 3Institute of Biology and Chemistry, Moscow Pedagogical State University, Moscow, Russia
- 4Tsitsin Main Botanical Garden, Russian Academy of Sciences, Moscow, Russia
- 5Faculty of Biology, Lomonosov Moscow State University, Moscow, Russia
- 6Moscow Institute of Physics and Technology, Moscow, Russia
Members of the genus Populus L. play an important role in the formation of forests in the northern hemisphere and are used in urban landscaping and timber production. Populus species of closely related sections show extensive hybridization. Therefore, the systematics of the genus is rather complicated, especially for poplars of hybrid origin. We aimed to assess the efficiency of application of the sex-determining region (SDR) in addition to the nuclear and chloroplast genome loci traditionally used in phylogenetic studies of poplars to investigate relationships in sections Aigeiros Duby and Tacamahaca Spach. Targeted deep sequencing of NTS 5S rDNA, ITS, DSH 2, DSH 5, DSH 8, DSH 12, DSH 29, 6, 15, 16, X18, trnG-psbK-psbI, rps2-rpoC2, rpoC2-rpoC1, as well as SDR and ARR17 gene was performed for 379 poplars. The SDR and ARR17 gene together with traditionally used multicopy and single-copy loci of nuclear and chloroplast DNA allowed us to obtain a clustering that is most consistent with poplar systematics based on morphological data and to shed light on several controversial hypotheses about the origin of the studied taxa (for example, the inexpediency of separating P. koreana, P. maximowiczii, and P. suaveolens into different species). We present a scheme of relationships between species and hybrids of sections Aigeiros and Tacamahaca based on molecular genetic, morphological, and geographical data. The geographical proximity of species and, therefore, the possibility of hybridization between them appear to be more important than the affiliation of species to the same section. We speculate that sections Aigeiros and Tacamahaca are distinguished primarily on an ecological principle (plain and mountain poplars) rather than on a genetic basis. Joint analysis of sequencing data for the SDR and chloroplast genome loci allowed us to determine the ancestors of P. × petrovskoe – P. laurifolia (female tree) × P. × canadensis (male tree), and P. × rasumovskoe – P. nigra (female tree) × P. suaveolens (male tree). Thus, the efficiency of using the SDR for the study of poplars of sections Aigeiros and Tacamahaca and the prospects of its use for the investigation of species of the genus Populus were shown.
1 Introduction
The genus Populus L. is distributed in the northern hemisphere from subtropical to boreal forests (DiFazio et al., 2011) and is one of the most economically and ecologically important genera of forest trees (Stettler et al., 1996). Poplar wood is used to produce plywood, pulp, paper, and biofuels (Kobolkuti et al., 2019). Species and interspecific hybrids of the genus Populus are widely used in landscaping cities and creating protective plantings because poplars have such valuable characteristics as fast growth, ecological plasticity, and ease of vegetative propagation (Bakulin, 2005). Poplars show high dust, smoke and gas resistance, and a huge leaf surface purifies the air of harmful impurities and absorbs a significant amount of CO2 (Kulagin, 1974; Jansson et al., 2010). In addition to their commercial value, poplars are ecologically important pioneers, colonizing disturbed and riverine ecosystems and contributing to the formation of natural forests in temperate and boreal regions, as well as in mountainous areas in the northern hemisphere (Schroeder et al., 2012).
The genus Populus, according to the Eckenwalder classification, includes 29 species in six sections: Abaso, Turanga, Populus (synonym: Leuce), Leucoides, Aigeiros, and Tacamahaca (Eckenwalder, 1996). Species of the genus Populus show extensive hybridization within sections as well as between closely related sections (Roe et al., 2014; Jiang et al., 2016). In this regard, the systematics of the genus Populus is rather complicated, and so far the researchers have not come to a consensus on the classification criteria, and the establishment of the taxonomic status of poplars of hybrid origin (both natural and cultivated) is particularly difficult (Cronk, 2005; Slavov and Zhelev, 2010; Schroeder et al., 2012; Mayorov et al., 2020). The question of the number of sections that should be distinguished in the genus Populus is also unresolved. For example, molecular genetic studies have shown that sections Leucoides, Aigeiros, and Tacamahaca should be considered not as separate sections but as a single subgenus, “Eupopulus” (Wang et al., 2020; Wang et al., 2022). Indeed, species of sections Aigeiros and Tacamahaca easily interbreed with each other, are connected by a powerful gene flow, and, in fact, have a common gene pool (at least, within Eurasia). Moreover, a common range of Eurasian poplars spreads from Western Europe to the Far East. Contacts between species in nature occur only at the borders of the habitats, i.e. “mixing” of genes occurs slowly. In this case, natural selection, despite gene flow, manages to maintain specificity of adaptive traits of each species, due to which it is possible to easily distinguish natural species morphologically (Nasimovich et al., 2019). In the study of cultivated poplars, identification is more difficult, but even in this case there is a factor supporting the discreteness of cultivars – the work of nurseries that propagate cultivars vegetatively. In this case, however, the control of varietal identity of seedlings is extremely important. When propagating poplars for use in urban landscaping, it is preferable to use only male clones that do not produce fluff (downy seeds) (Melnikova et al., 2019; Huang et al., 2022). However, it is impossible to determine the sex of a 1-2-year-old seedling by its phenotype; this can only be done after a few years when an adult tree begins to flower. In the cities of Central Russia, poplar was actively used in landscaping, but many female clones were mistakenly planted, which leads to annual problems with poplar fluff, because it collects and carries allergens, clogs drains and ventilation systems, is a fire hazard, and so on (Janković et al., 2021; Melnikova and Dmitriev, 2022). Genetic testing of seedlings to determine the sex would help not to repeat previous mistakes. In addition, when creating poplar hybrids for urban landscaping or industrial wood use, it is advisable to properly select parental species considering their adaptation to certain habitats (for example, representatives of the section Aigeiros are mostly plain poplars, and the section Tacamahaca includes mostly mountain poplars) (Nasimovich et al., 2019). Thus, understanding the evolutionary relationships of Populus species is important for basic research and breeding work (Wang et al., 2014).
A significant number of studies have been devoted to the search for reliable molecular markers for establishing phylogenetic relationships between poplar species – an analysis of various regions of nuclear DNA as well as sequences of the chloroplast genome was carried out (Schroeder et al., 2012; Wang et al., 2014; Alexandrov and Karlov, 2018; Zhou et al., 2018; Wang et al., 2022). The informativity for phylogenetic studies of poplars was noted for the sequences of non-transcribed intergenic spacers (NTS) of the 5S ribosomal RNA genes (rDNA) (Negi et al., 2002; Wilson, 2013; Alexandrov and Karlov, 2018) and internal transcribed spacers of rRNA genes (ITS) (Leskinen and Alström-Rapaport, 1999). At the same time, there are discrepancies between the phylogenetic trees obtained by analyzing the chloroplast and nuclear genome sequences of Populus species, which are usually attributed to their polyphyletic or hybrid origins (Hamzeh and Dayanandan, 2004; Wang et al., 2014; Liu et al., 2016; Zhou et al., 2018; Wang et al., 2022). In the analysis of chloroplast genome sequences, some accessions of the same species (a prime example is P. nigra) did not cluster together, but there was a significant association with geographic origin, which can be explained by recent hybridization (Wang et al., 2022). Analysis of different sequences allows clarification of various issues: chloroplast genome sequences showed great informativity for determining interspecific relationships in the genus Populus, including taxa difficult to systematize, while nuclear genome sequences were more effective for studying the deep phylogeny of Populus (Wang et al., 2022).
Poplars are dioecious plants, and another promising sequence for clarifying the systematic relationships of species and hybrids of the genus Populus is the sex-determining region (SDR). Sex chromosomes are actively used in phylogenetic studies (Parker et al., 2020; Navarro-Lopez et al., 2021). The species of sections Aigeiros and Tacamahaca are characterized by an XY sex-determination system (Gaudet et al., 2008; Pakull et al., 2009; Pakull et al., 2011; Kersten et al., 2014; Geraldes et al., 2015; Pakull et al., 2015; McKown et al., 2017), which allows the examination of poplar relationships in the male lineage. A series of studies made it possible to identify sex-associated DNA polymorphisms, determine the location and sequence of the SDR in various species of the genus Populus, and establish that sex is determined by the ARABIDOPSIS RESPONSE REGLATOR 17 (ARR17) gene – when ARR17 is turned on, female flowers develop, and when it is turned off – men ones (Gaudet et al., 2008; Yin et al., 2008; Pakull et al., 2009; Paolucci et al., 2010; Kersten et al., 2014; Geraldes et al., 2015; Pakull et al., 2015; McKown et al., 2017; Melnikova et al., 2019; Muller et al., 2020; Xue et al., 2020; Zhou et al., 2020; Kim et al., 2021; Melnikova et al., 2021; Yang et al., 2021). In species of sections Aigeiros and Tacamahaca, the SDR is localized on the end of chromosome 19, has a length of about 120 thousand nucleotides, and contains several genes, including T-complex protein 1 subunit gamma (TCP), Chloride channel protein CLC-c (CLC), and DNA-methyltransferase 1 (MET1). Only in the SDR of male plants, there are partial repeats of the ARR17 gene, from which small RNAs are synthesized, leading to the inactivation of ARR17, which is localized on the opposite to the SDR end of chromosome 19 (Xue et al., 2020; Zhou et al., 2020; Melnikova et al., 2021). Although the ARR17 gene plays a key role in sex determination in the genus Populus, species of this genus differ in the sex-determination system and structure of the SDR. P. euphratica and P. tremula have an XY sex-determination system, however, in P. tremula, the SDR containing partial ARR17 repeats is located in the peritelomeric region of chromosome 19, while in P. euphratica, it is located in the peritelomeric region of chromosome 14. Moreover, the SDR of these species is several hundred thousand nucleotides, differing significantly from the SDR of poplars of sections Aigeiros and Tacamahaca (Muller et al., 2020; Zhang et al., 2022). In P. alba, three copies of the ARR17 gene are located in the peritelomeric region of chromosome 19 in female plants, whereas in male plants, ARR17 is absent, indicating that P. alba has an ZW sex-determination system (Muller et al., 2020).
Previously, we showed that SDR polymorphisms can be effectively used in assessing the genetic proximity of Aigeiros and Tacamahaca species and hybrids based on whole genome sequencing data (Melnikova et al., 2021). The aim of this work was to evaluate the efficiency of using the SDR to establish the relationships of poplar species and hybrids of sections Aigeiros and Tacamahaca in combination with the nuclear and chloroplast genome sequences that have proven themselves in the study of the genus Populus systematics based on the target deep sequencing data.
2 Materials and methods
2.1 Plant material
The first part of the collection was formed from 222 samples of poplars of sections Aigeiros and Tacamahaca collected on the territory of Moscow (Russia). Plant material was gathered in different areas of the city to maximize the coverage of the existing genetic diversity. Samples were collected during flowering of poplar trees for accurate sex determination. The second part of the collection was formed from 157 samples of sections Aigeiros and Tacamahaca; the material was collected in 15 different regions of Russia: Moscow and Moscow Region, Orenburg Region, Novosibirsk Region, Altai Republic, Kemerovo Region, Republic of Khakassia, Krasnoyarsk Territory, Transbaikal Territory, Republic of Sakha (Yakutia), Jewish Autonomous Region, Khabarovsk Territory, Primorsky Territory, Sakhalin Region, and Magadan Region, as well as in natural areas of poplar species in 5 countries: Italy, Uzbekistan, Kazakhstan, Kyrgyzstan (Irkeshtam), Mongolia. Part of the accessions were herbarium samples from the collection of the Tsitsin Main Botanical Garden, Russian Academy of Sciences (MHA, Moscow, Russia). The resulting collection consisted of 379 accessions of poplars of sections Aigeiros and Tacamahaca. Affiliation of a poplar to a particular species/hybrid was carried out using a key for determining poplars (Mayorov et al., 2020). A complete list of accessions is given in Supplementary Data 1, and a description of poplar species and hybrids can be found in Supplementary Data 2.
Accessions from the section Tacamahaca were represented by the following species: P. suaveolens – 38 samples, P. laurifolia – 18 samples, P. talassica – 10 samples, P. longifolia – 6 samples (and 3 more samples with doubts in identification), P. maximowiczii – 8 samples, P. simonii – 8 samples, P. koreana – 5 samples, and P. trichocarpa – 4 samples. Accessions from the section Aigeiros were represented by the following species: P. nigra – 22 samples, P. nigra × P. pyramidalis – 3 samples, P. afghanica – 3 samples, and P. deltoides – 1 sample. The sample set also included the hybrids most commonly used in Moscow landscaping: P. × sibirica – 34 samples (and 27 more samples with doubts in identification), P. × petrovskoe – 57 samples (and 5 more samples with doubts in identification), P. × rasumovskoe – 30 samples (and 4 more samples with doubts in identification), P. × canadensis – 3 samples, as well as a rarely cultivated hybrid P. × wobstii (3 samples) and a natural Siberian hybrid P. × irtyschensis (7 samples). We also studied P. nigra × P. × sibirica – 8 samples, P. deltoides × (P. laurifolia × P. suaveolens) – 4 samples, P. deltoides × P. suaveolens – 1 sample. In 5 cases, more complex hybrids were examined. Five samples could not be identified reliably, and two possible affiliations were given for them. For 57 samples, the species affiliation was not established.
2.2 Description of the studied species and hybrids of poplar
The following is an enumeration of the studied taxa, along with a list of diagnostic features for poorly known taxa. We described in detail hybrid taxa for which mutually exclusive data are available in the literature on diagnostic traits and parental species. In these cases, we have to give detailed descriptions so that it is clear exactly what our genetic data refer to. For more details, see the full description of the studied taxa in Supplementary Data 2.
2.2.1 Section Aigeiros Duby
Mainly plain poplars; petioles are long, strongly flattened laterally, without an adaxial groove, usually glabrous; leaf blades are rhombic or deltate, but, in some species, with rounded lateral edges; the maximum expansion of a leaf blade is very strongly shifted toward the base.
P. afghanica C.K.Schneid.
Crown is spreading or pyramidal
Petioles are slightly pubescent or glabrous
Leaf blades are rhombic-like or deltate, but with rounded lateral edges
Note: morphologically close to P. nigra; however, it has certain features of “balsam” poplars
P. × canadensis Moench [P. deltoides × P. nigra; cultivar]
Crown is spreading
Petioles are glabrous
Glands occur only on half of the leaves
Leaf blades are deltate or rhombic-like
P. deltoides W.Bartram ex Marshall
Crown is spreading
Petioles are glabrous
Leaf blades are deltate
Glands are on most leaves
Note: leaves are particularly large
P. nigra L. (P. nigra var. nigra)
Crown is spreading
Petioles are glabrous
Leaf blades are rhombic-like, less often deltate
Glands are usually absent or small
P. nigra × P. pyramidalis Rozier (P. nigra var. nigra × P. nigra var. italica Munchh.; cultivar).
Crown is pyramidal
Petioles are slightly pubescent, less often glabrous
Leaf blades are rhombic-like, less often deltate, with slightly rounded edges
Glands are usually small or absent
Note: mixes with more southern P. pyramidalis Rozier; represented in the dendrograms under this name
2.2.2 Section Tacamahaca Spach
Predominantly mountain poplars; petioles are terete (rounded in cross section) and shorter on average than those of “balsam” poplars, usually pubescent, with an adaxial groove; leaf blades are of various shapes (lanceolate, ovate, slightly cordate, etc.), but not rhombic-like or deltate; the maximum expansion of a leaf blade is in its middle or shifted toward base or apex.
P. koreana Rehder
Axes of 1-2-year-old shoots are terete
Leaf blades are angular (not flat), matte, strongly leathery
Leaf blade apex is acuminate, tapering to a tip up to 5-6 mm long
Midrib is pubescent on the abaxial side
Capsules have 3 glabrous valves
P. laurifolia Ledeb.
Axes of 1-2-year-old shoots are strongly grooved
Leaf blades are flat, matte, not leathery
Leaf blade apex is acute or attenuate, without a long tip
Midrib is glabrous
Capsules usually have 3 glabrous valves
P. longifolia Fisch.
Axes of 1-2-year-old shoots are terete
Leaf blades are flat, matte, not leathery
Leaf blade apex is acute or attenuate, without a long tip
Midrib is glabrous
Capsules have 3 glabrous valves
Note: a medium-height tree; forms abundant sucker shoots; the color contrast of the adaxial and abaxial sides of leaf blades is maximal among the section Tacamahaca representatives; petioles are long
P. maximowiczii Henry
Axes of 1-2-year-old shoots are terete
Leaf blades are flat, slightly glossy, slightly leathery
Leaf blade apex is acuminate, tapering to a tip up to 5-6 mm long
Midrib is pubescent on the adaxial and abaxial sides
Capsules have 3 glabrous valves
P. simonii Carriere f. fastigiata
Axes of 1-2-year-old shoots are strongly grooved
Leaf blades are flat, matte, not leathery
Leaf blade apex is acute or obtuse, sometimes apiculate, with a 0.5-3 mm tip
Midrib is glabrous
Note: crown is pyramidal or wide pyramidal; leaf blades are obovate, always with attenuate base
P. simonii Carriere f. subpendula
Leaf blades are flat, matte, not leathery
Leaf blade apex is acute, sometimes attenuate, without a long tip
Midrib is glabrous
Note: crown is semi-weeping; leaf blades are lanceolate (cuneate base and acute apex), sit on short shoots (brachyblasts)
P. suaveolens Fisch.
Axes of 1-2-year-old shoots are terete
Leaf blades are flat, matte, not leathery
Leaf blade apex is acuminate, tapering to a tip up to 5-6 mm long
Midrib is glabrous
Capsules have 3 glabrous valves
P. talassica Kom.
Axes of 1-2-year-old shoots are angular
Leaf blades are flat, matte, not leathery
Leaf blade apex is acute or attenuate
Midrib is glabrous
Capsules have 3 glabrous valves
? P. trichocarpa Torr. et A.Gray ex Hook. [? P. × trichocarpa ‘Lettland’]
Here and further the sign «?» before a species/hybrid means that we have doubts in accurate identification of species affiliation
Axes of 1-2-year-old shoots are terete or grooved
Leaf blades are flat, matte, not leathery
Leaf blade apex is acute, without a narrow tip
Capsules have 3 pubescent valves
P. × wobstii R.I.Schrod. (P. laurifolia and P. longifolia)
Axes of 1-2-year-old shoots are slightly grooved
Leaf blades are flat, matte, not leathery
Leaf blade apex is acute, without a narrow tip
Midrib is glabrous
Capsules have 3 glabrous valves
2.2.3 Intersectional hybrids Aigeiros × Tacamahaca
This group includes the most widespread poplar hybrids (hybridogenic species, spontaneous hybrids, cultivars) of urban landscaping in Russia. Some of them are known under erroneous names in Russia and sometimes beyond its borders. These names may not coincide in Russia and other countries and may be different in the understanding of different authors within the same country. Therefore, we provide the main diagnostic features of these hybrids and sometimes complete descriptions (see also Supplementary Data 2).
The common features of these poplars are the following: petioles are long or medium, slightly flattened laterally, with a narrow and interrupted adaxial groove, usually slightly pubescent; leaf blades have diverse forms (lanceolate, ovate, cordate, etc., often rhombic-like); the maximum expansion of a leaf blade is strongly shifted toward the base.
P. deltoides × (P. laurifolia × P. suaveolens)
Crown is spreading
Axes of 1-2-year-old shoots are terete, angular, sometimes slightly grooved
Leaf blades are ovate or cordate, the length exceeds the width by 1.5 times on average
Leaf blade base is rounded, cuneate, or cordate
Leaf blade apex is attenuate
Capsules have 3 valves
P. × irtyschensis Chang Y.Yang [P. × berolinensis nothovar. irtyschensis (Chang Y.Yang) C.Shang; P. laurifolia × P. nigra var. nigra]
Crown is spreading and irregular
Axes of 1-2-year-old shoots are angular or slightly grooved
Leaf blades are ovate-rhombic; the length exceeds the width by 1.5 times on average
Leaf blade base is rounded, cuneate, or irregular as in P. × sibirica
Leaf blade apex is acute or attenuate
Capsules have 2-3 valves
P. × petrovskoe R.I.Schrod. ex Wolkenst. [In Russia, it is mentioned under the erroneous name of P. × berolinensis K.Koch; ? P. × canadensis × P. laurifolia]
Crown is spreading, regular, wide, becomes semi-pyramidal with age
Axes of 1-2-year-old shoots are angular or grooved
Leaf blades are ovate-rhombic, the length exceeds the width by 1.2 times on average
Leaf blade base is irregular: rounded at the petiole and broadly cuneate at a distance from it
Leaf blade apex is attenuate
Capsules have 3 valves
P. × rasumovskoe R.I.Schrod. ex Wolkenst. [P. nigra var. nigra × P. suaveolens]
Crown is spreading, irregular, usually with hanging branch ends
Axes of 1-2-year-old shoots are terete
Leaf blades are lanceolate or ovate
Leaf blade base is rounded
Leaf blade apex is acuminate with a 1-1.5 cm tip
Note: the maximum expansion of a leaf blade is sometimes shifted toward the base because of the long tip
P. × sibirica G.V.Krylov et G.V.Grig. ex A.K.Skvortsov [? P. nigra var. nigra × (P. laurifolia × P. suaveolens)]
Crown is spreading, irregular
Axes of 1-2-year-old shoots are terete or angular, rarely slightly grooved
Leaf blades are ovate-rhombic, the length exceeds the width by 1.5-2 times on average
Leaf blade base is irregular: rounded at the petiole and cuneate at a distance from it
Leaf blade apex is attenuate (1-2 cm tip)
Capsules have 2 or 2-3 valves
Note: leaves turn yellow in autumn and fall off earlier than in other analyzed intersectional poplar hybrids
2.3 DNA isolation for sequencing on the Illumina platform
For the accessions collected in Moscow, the 0.2 g leaf material was homogenized using a MagNA Lyser automatic homogenizer (Roche, Switzerland) in 1 ml of lysing modified CTAB buffer (100 mM Tris-HCl pH 8.0 (VWR Life Science, USA); 3% CTAB (VWR Life Science); 3M NaCl (Scharlab, Spain); 20 mM EDTA pH 8.0 (Promega, USA); 1% PVP K30 (PanReac AppliChem)) with 5 μl β-mercaptoethanol (BioRad, USA) and Solid-glass beads (Sigma-Aldrich, USA). The homogenate was incubated in a Gnom thermostat (DNA-Technology, Russia) at 65°C for 1 h, stirring every 20 min. Next, two consecutive purifications were performed with chloroform: an equal volume of chloroform (Acros Organics, USA) was added to the homogenate, and then centrifuged on a 5418R microcentrifuge (Eppendorf, Germany) for 10 min at 10,000 g and 4°C. The aqueous phase was then transferred to clean tubes, and two volumes of 96% alcohol were added to precipitate the DNA and incubated for 30 min at -20°C. Then, it was centrifuged for 15 min at 10,000 g and 4°C. After that, the supernatant was carefully removed without touching the precipitate. Next, the precipitate was washed with 70% alcohol and centrifuged for 5 min at 10,000 g and 4°C. The DNA precipitate was air dried for 5 min and dissolved in 100 µl of the DNA dilution buffer (Evrogen, Russia). The DNA concentration was estimated using a Qubit fluorometer (Thermo Fisher Scientific, USA). For the accessions provided from different regions of Russia, DNA was isolated with the classical CTAB method (Doyle and Doyle, 1987).
2.4 Preparation of DNA libraries for targeted deep sequencing on the Illumina platform
Preparation of DNA libraries included amplification of target nucleotide sequences and adding to each amplicon indexes unique to a sample and adapters required for sequencing. We designed primers for amplification of the SDR and ARR17 gene loci, as well as applied primers previously used for amplification of single-copy nuclear DNA genes, namely DSH 2, DSH 8, DSH 29, 6, 15, 16, X18, DSH 5, DSH 12 (Du et al., 2014; Wang et al., 2014; Wang et al., 2019), chloroplast genome loci, namely trnG‐psbK-psbI, rps2‐rpoC2, rpoC2‐rpoC1 (Schroeder et al., 2012; Huang et al., 2014; Zhou et al., 2018), and ITS (Hsiao et al., 1994) and NTS 5S rDNA (Falistocco et al., 2007). All primers were extended with sequences required for sample preparation and sequencing of DNA libraries on the Illumina platform as described in our previous works (Melnikova et al., 2019; Dmitriev et al., 2020; Novakovskiy et al., 2020; Povkhova et al., 2022) (a table with primers and a figure with the location of the amplified fragments in the SDR and ARR17 gene are represented in Supplementary Data 3). Briefly, two sequential PCR were used to prepare DNA libraries: the first to amplify the target genome regions and add universal sequences to them (specific primers were used) and the second to add the sequences required for sequencing and double-indexes (Nextera XT v2 Illumina universal primers were used) as described previously (Melnikova et al., 2019; Dmitriev et al., 2020; Novakovskiy et al., 2020; Povkhova et al., 2022). The concentration and quality of the prepared DNA libraries were assessed using a Qubit fluorometer (Thermo Fisher Scientific) and an Agilent 2100 bioanalyzer (Agilent Technologies, USA). The obtained DNA libraries were diluted to a concentration of 4 nM. Sequencing of DNA libraries was performed on a MiSeq instrument (Illumina, USA) using the MiSeq Reagent Kit v3 (600 cycles) with reads of 300 nucleotides on each side.
2.5 Bioinformatics data analysis
The reads were trimmed from the 3’-end by quality, and adapter sequences were removed using Trimmomatic 0.39. The processed reads were then mapped to the regions of the reference genome of the male P. trichocarpa “Stettler 14” v1.1 (https://phytozome-next.jgi.doe.gov/info/PtrichocarpaStettler14 v1 1, (Hofmeister et al., 2020)) corresponding to the sequenced loci using BWA 0.7.17 with parameters for increased sensitivity due to probably low homology with the reference for some species/hybrids (Li, 2013). Next, the obtained BAM files were sorted using Samtools 1.10. The search for single-nucleotide polymorphisms (SNPs) and short insertions/deletions (InDels) was performed using freeBayes 1.3.2 (Garrison and Marth, 2012). Then, from the VCF files, the frequencies of SNP allele variants (variant allele frequency, VAF) were determined in different samples and pairwise Euclidean distances between samples were calculated based on the obtained VAF vectors. Clustering was performed with Ward’s method (ward.D2) in R 4.2.1. Visualization was performed using the ggtree and ggplot2 packages for R. Bootstrap analysis was performed using pvclust (Suzuki and Shimodaira, 2006). In our dataset, the majority of samples are hybrids of different species of sections Aigeiros and Tacamahaca (from 2 to 4-5 different parental species). In bootstrap analysis, they jump from cluster to cluster during subsampling because they have genetic similarity to different parental species. Thus, the bootstrapping method is impractical here to assess the reliability of the results. In this respect, it is better to consider the genetic distance matrices (calculated based on VAF) on the basis of which the clustering was performed.
3 Results
3.1 Results of targeted deep sequencing of 379 poplar accessions
For 379 poplar accessions of sections Aigeiros and Tacamahaca, 14 loci previously used in phylogenetic studies of poplars were sequenced, including NTS 5S rDNA, ITS, DSH 2, DSH 5, DSH 8, DSH 12, DSH 29, 6, 15, 16, X18, trnG-psbK-psbI, rps2-rpoC2, rpoC2-rpoC1. In addition, loci of the sex-determining region (SDR) and ARR17 gene, which determines sex in the genus Populus and whose partial repeats are located in the SDR, were sequenced. An average of about 500 reads were obtained for each sequenced amplicon of each sample. A search for polymorphisms was performed based on the sequencing data. The results are represented in Supplementary Data 4. We also evaluated the genetic proximity of the studied poplar accessions based on the identified polymorphisms. The results are represented in Supplementary Data 5. To visualize the data obtained on the genetic proximity of the studied poplar accessions, dendrograms were constructed based on the polymorphisms of the individual studied loci, as well as their combinations.
3.2 Analysis of 14 loci traditionally used in poplar phylogenetic studies
Figure 1 (Supplementary Data 6A, 8A) shows the results of clustering poplar samples based on sequencing data for 14 loci previously used in phylogenetic studies of Populus species: NTS 5S rDNA, ITS, DSH 2, DSH 5, DSH 8, DSH 12, DSH 29, 6, 15, 16, X18, trnG‐psbK-psbI, rps2‐rpoC2, rpoC2-rpoC1 (White et al., 1990; Schmidt et al., 1994; Hsiao et al., 1995; Schroeder et al., 2012; Huang et al., 2014; Wang et al., 2014; Zhou et al., 2018; Wang et al., 2019). On the outer circle, the species affiliations of the studied 379 poplar accessions are marked with different colors. On the middle circle, blue and green colors indicate male and female genotypes, respectively. On the inner circle, data on read coverage for each examined sample are shown in the form of a heat scale.
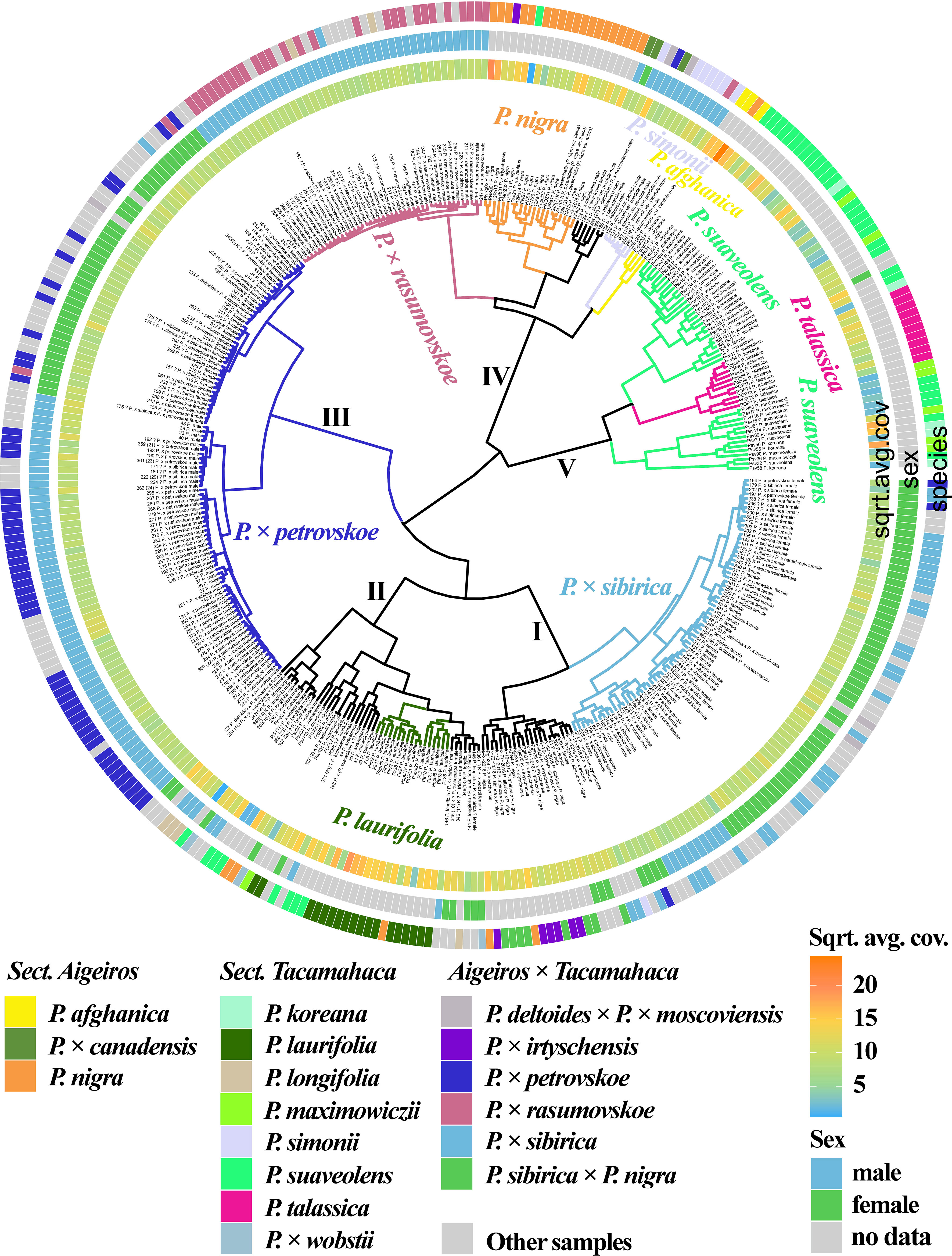
Figure 1 Dendrogram based on deep sequencing data for NTS 5S rDNA, ITS, DSH 2, DSH 5, DSH 8, DSH 12, DSH 29, 6, 15, 16, X18, trnG‐psbK-psbI, rps2-rpoC2, and rpoC2-rpoC1 sequences. Colors corresponding to species and hybrids mark only accessions for which there were no doubts in the morphological determination of the species affiliation.
A large cluster (I) was identified that included predominantly male and female samples of P. × sibirica, as well as hybrids in which one of the putative parental forms was P. × sibirica, all but one samples of P. × irtyschensis, and two samples of P. nigra (KPN4 and k-2-216). Another cluster (II) included predominantly male and female accessions of P. laurifolia, P. longifolia, some of P. suaveolens, P. × wobstii, and a few P. nigra. One more cluster (III) included the vast majority of P. × petrovskoe accessions, which were divided into two subclusters according to sex (male or female). The next cluster (IV) included almost all accessions of P. × rasumovskoe, forming a separate subcluster, and subclusters of P. nigra, P. simonii, and P. afghanica, as well as P. deltoides and its hybrids (including P. × canadensis). The latter cluster (V) combined subclusters of P. suaveolens, P. koreana, P. maximowiczii, and P. talassica. In all clusters, in addition to the listed accessions, there were also samples for which there were doubts in the accuracy of species identification (marked “?”) and samples for which determination of species affiliation on the basis of morphological analysis was not carried out.
The resulting dendrogram divided the samples quite well according to their species affiliation – thus, three clear clusters of hybrids (P. × sibirica, P. × petrovskoe, P. × rasumovskoe) and two clusters of “pure” or hybridogenic species were distinguished. The distinguished subclusters also corresponded to the division of samples according to the species affiliation. In addition, genetically related accessions were combined into clusters. For example, the P. × rasumovskoe cluster included most accessions of its probable parental form, P. nigra. P. laurifolia, which can be considered as a dynamic state of P. suaveolens under conditions of continuous gene flow from P. nigra (Nasimovich et al., 2019), formed one cluster with some of P. suaveolens. However, although we mostly saw a good division of accessions into small subclusters according to their species affiliation, a cluster could include several small clusters of accessions of different species interspersed with each other, as can be clearly seen for cluster II.
We also analyzed the dendrograms obtained from the sequencing data separately for each of the 14 studied loci and compared them with the dendrogram obtained from the data for all 14 loci. Low read coverage was observed for a number of samples for some sequenced loci (such samples are marked in dark blue in the heat scale of the inner circle); we do not draw any conclusions for them in the analysis described below.
3.2.1 Analysis of multicopy loci of nuclear genome (NTS 5S rDNA, ITS)
We analyzed the dendrogram obtained on the basis of NTS 5S rDNA (chromosome 17) (Xin et al., 2020) (Supplementary Data 6B, 8B), which showed efficiency in phylogenetic studies of poplars (Negi et al., 2002; Wilson, 2013; Alexandrov and Karlov, 2018). Samples of P. × sibirica, P. × petrovskoe, and P. × rasumovskoe formed separate groups. The P. × sibirica and P. × rasumovskoe subclusters merged into one cluster with the group of P. suaveolens samples, which is their probable ancestor. However, for many “pure” and hybridogenic species, the picture was ambiguous – they were all included in the same cluster, but did not form clear subclusters corresponding to individual species, like in the dendrogram based on 14 loci, although regularities corresponding to the phylogenetic relationships of poplars can be traced.
For another sequence frequently used in plant phylogenetic studies, ITS (chromosomes 8 and 14) (Xin et al., 2020), the clustering results were less consistent with the known information on poplar systematics and were inferior to NTS 5S rDNA in informativity (Supplementary Data 6C, 8C). Thus, groups of P. × petrovskoe and P. × rasumovskoe were identified, but for P. × sibirica some samples were not in the main cluster of this hybrid, but in a cluster with different species. A number of accessions were grouped according to the species affiliation (e.g., the P. simonii subcluster), however, many samples of different species did not form separate subclusters but were interspersed with accessions of other species.
3.2.2 Analysis of single-copy loci of nuclear genome (DSH 2, DSH 5, DSH 8, DSH 12, DSH 29, 6, 15, 16, X18)
The DSH 2 locus (chromosome 4) (Du et al., 2014) was sufficiently informative (Supplementary Data 6D, 8D). Its analysis identified subclusters of P. × rasumovskoe, predominantly female samples of P. × sibirica, male samples of P. × petrovskoe, and female samples of P. × petrovskoe. However, female accessions of P. × petrovskoe were closer to female accessions of P. × sibirica than to male accessions of P. × petrovskoe. Some of the male samples of P. × sibirica clustered with the female samples of this hybrid, while others formed a separate distant subcluster. Subclusters of mainly samples of P. nigra, P. talassica, P. laurifolia, P. simonii, or P. suaveolens (together with P. koreana and P. maximowiczii, which can be considered as P. suaveolens) were also identified. However, some of the samples, like in the NTS 5S rDNA and ITS analyses, did not fall into the subclusters of the species to which they belong, but found themselves in other subclusters. In addition, subclusters were formed from samples of different species.
Analysis based on the DSH 8 locus (chromosome 18) (Du et al., 2014) allowed us to distinguish a number of sample groups according to their taxonomic affiliation (Supplementary Data 6E, 8E). Subclusters comprising predominantly accessions of one species/hybrid were identified for P. × rasumovskoe, P. × sibirica, P. × petrovskoe, P. nigra, and P. simonii. However, often, individual samples of the same species were not grouped together, but turned out to be in remote clusters. A striking example of such a pattern is P. talassica, which divided into groups.
When analyzed by the DSH 29 locus (chromosome 14) (Wang et al., 2014), subclusters of mostly samples of the same species/hybrid were formed for P. × rasumovskoe, P. × petrovskoe (male and female plants were divided into two subclusters, the male subcluster also included samples of P. × sibirica male plants), P. nigra, and P. × sibirica (female plants) (Supplementary Data 6F, 8F). For most other species, however, the division into groups according to taxonomic affiliation was unclear; species from different sections were grouped in the same cluster. For example, P. simonii (Tacamahaca) samples were in a subcluster with the majority of P. nigra (Aigeiros) samples; samples of P. laurifolia, P. talassica, P. suaveolens (together with P. koreana and P. maximowiczii) (Tacamahaca) and P. nigra (Aigeiros) formed mixed subclusters.
The analysis for gene 6 (chromosome 12) (Wang et al., 2019) identified clusters of the majority of samples of P. × rasumovskoe, P. × petrovskoe (divided into two subclusters according to sex), or P. × sibirica (Supplementary Data 6G, 8G). However, separation of samples into subclusters corresponding to species was revealed only for small groups of accessions; there was no clear division according to taxonomic affiliation.
The gene 15 (chromosome 15) (Wang et al., 2019) analysis identified a cluster of P. × rasumovskoe, while P. × petrovskoe and P. × sibirica samples formed a mixed cluster (Supplementary Data 6H, 8H). In addition, a cluster of the majority of P. nigra samples was identified, but several P. nigra samples were in other clusters. Samples of most species formed mixed clusters, which in some cases had isolated subclusters consisting of a small number of accessions of one species. Species from sections Aigeiros and Tacamahaca were often grouped in the same cluster.
The analysis of gene 16 (chromosome 3) (Wang et al., 2019) allowed us to distinguish subclusters of mainly P. × rasumovskoe or P. × petrovskoe (male trees) samples (Supplementary Data 6I, 8I). A subcluster of the majority of P. nigra samples was identified, which also included P. × irtyschensis and P. × canadensis accessions. A subcluster of P. simonii samples was also distinguished. In addition, a cluster of a mixture of P. × petrovskoe and P. × sibirica female accessions was revealed. In some cases (e.g., for P. talassica, P. suaveolens (together with P. koreana and P. maximowiczii)), there were isolated small subclusters of samples of the same species, but often accessions of different species formed mixed subclusters.
The analysis of the X18 locus (chromosome 15) (Wang et al., 2019) identified a cluster of P. × petrovskoe with a division into male and female genotypes (Supplementary Data 6J, 8J). This cluster also included subclusters of P. nigra samples, which is the putative ancestor of P. × petrovskoe. Samples of P. × rasumovskoe and P. × sibirica were in one cluster and appeared to be mixed with each other. A subcluster of P. simonii, several subclusters of P. suaveolens, P. nigra, and P. laurifolia were identified, but a significant number of subclusters consisted of a mixture of samples of different taxa, including species from different sections.
The analyses based on DSH 5 (chromosome 12) (Du et al., 2014) and DSH 12 (chromosome 2) (Du et al., 2014) were low-informative (Supplementary Data 6K, L, 8K, L). The division of samples into clusters/subclusters according to taxonomic affiliation was poorly pronounced.
3.2.3 Joint analysis of 11 loci of nuclear genome
Based on the analysis of the NTS 5S rDNA, ITS, DSH 2, DSH 5, DSH 8, DSH 12, DSH 29, 6, 15, 16, X18 sequences (Figure 2, Supplementary Data 6M, 8M), the P. × petrovskoe cluster (I) was isolated, with the samples divided into two subclusters according to sex. The P. × rasumovskoe subcluster was also distinguished and formed one cluster (II) with the subcluster of P. simonii. Samples of P. × sibirica formed a separate subcluster. A subcluster of the vast majority of P. nigra and P. × irtyschensis samples, all samples of P. nigra × P. × sibirica and P. × canadensis, and a single sample of P. deltoides merged into one cluster (III) with the P. × sibirica subcluster. Samples of P. suaveolens and P. laurifolia were found in two clusters (IV and V) and formed subclusters or interspersed with each other or with accessions of other species. P. longifolia, P. afghanica, and P. × wobstii were in cluster V, while P. talassica samples were in cluster IV. Clusters IV and V also contained one P. nigra accession. Thus, samples of different species were combined into small groups, but groups of different species alternated with each other – no large isolated subclusters were formed according to taxonomic affiliation.
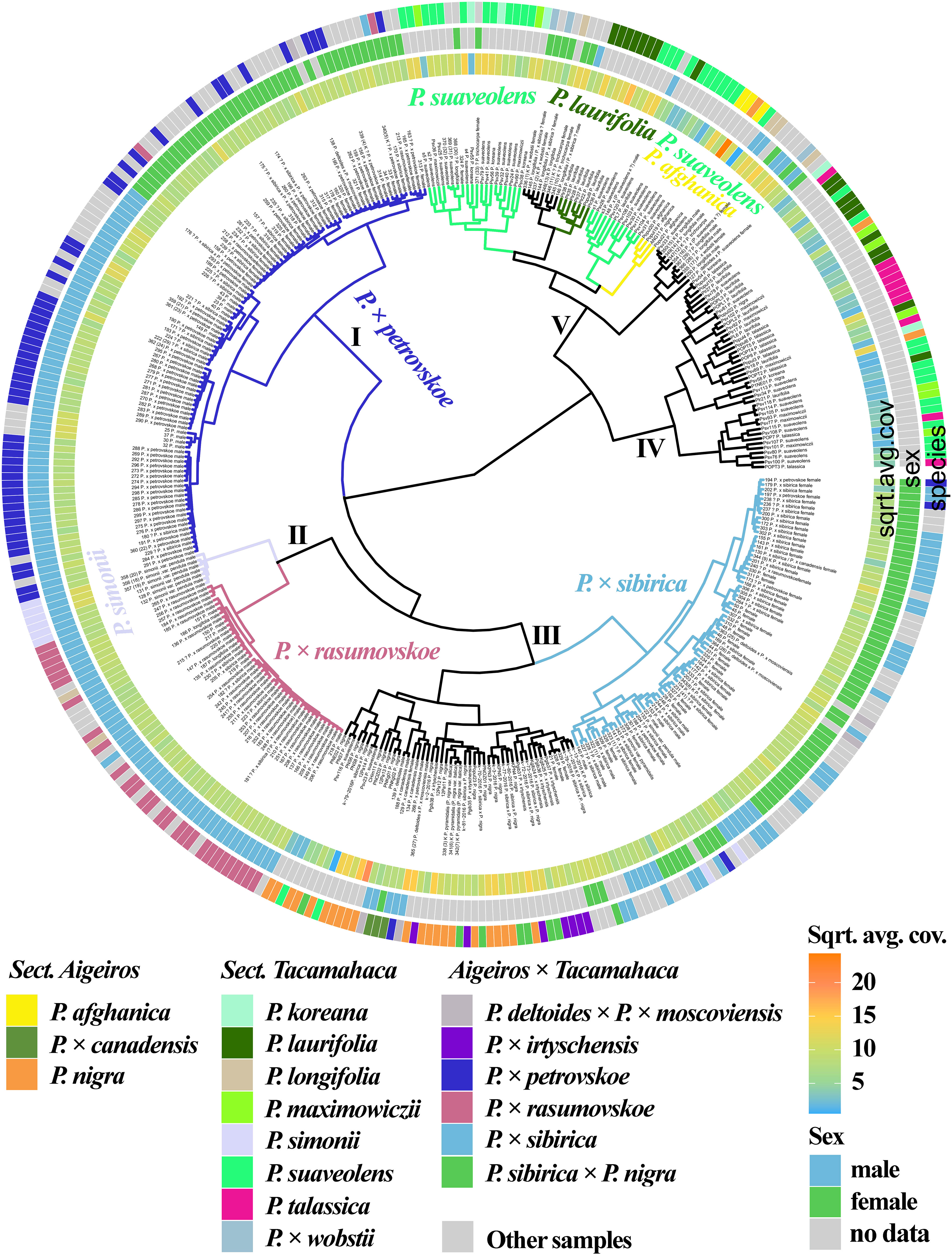
Figure 2 Dendrogram based on deep sequencing data for NTS 5S rDNA, ITS, DSH 2, DSH 5, DSH 8, DSH 12, DSH 29, 6, 15, 16, and X18 sequences. Colors corresponding to species and hybrids mark only accessions for which there were no doubts in the morphological determination of the species affiliation.
3.2.4 Analysis of chloroplast loci (trnG‐psbK-psbI, rps2‐rpoC2, rpoC2-rpoC1)
Based on the trnG‐psbK-psbI (Schroeder et al., 2012) analysis (Supplementary Data 7A, 8N), P. × rasumovskoe samples were separated into a distinct cluster with the majority of P. nigra samples, which is considered one of the probable parental species of P. × rasumovskoe (Mayorov et al., 2020). The cluster of the majority of P. × petrovskoe samples also included a number of samples of P. laurifolia, a probable parental species of P. × petrovskoe (Mayorov et al., 2020). P. talassica, P. afghanica, P. simonii, and P. suaveolens formed subclusters (several groups) according to the species affiliation. Samples of P. × sibirica did not form a separate cluster but mixed with samples of other species, including P. longifolia, P. suaveolens, P. laurifolia, P. × wobstii, P. × irtyschensis, and several samples of P. nigra. The trnG-psbK-psbI sequence proved to be the most informative of the chloroplast loci we studied.
The rps2‐rpoC2 (Schroeder et al., 2012) analysis (Supplementary Data 7B, 8O) isolated the P. × rasumovskoe cluster, which also included most P. nigra samples. Subclusters of P. suaveolens (together with P. koreana and P. maximowiczii), P. talassica, and P. simonii were distinguished. No isolated clusters were identified for P. × petrovskoe and P. × sibirica; these accessions alternated with each other as well as with accessions of P. laurifolia, P. × wobstii, and P. × irtyschensis.
Based on the rpoC2‐rpoC1 (Schroeder et al., 2012) analysis (Supplementary Data 7C, 8P), a cluster of P. × rasumovskoe was identified, which also contained several samples of P. nigra and single accessions of other species. P. × petrovskoe accessions formed a large cluster, in which they were interspersed with samples of P. × sibirica, as well as samples of many other studied species, without forming subclusters according to taxonomic affiliation. In most cases, there was no grouping of accessions according to belonging to a particular species and section. This locus proved to be of little informative value.
The results based on the analysis of three chloroplast genome loci (trnG-psbK-psbI, rps2-rpoC2, rpoC2-rpoC1) simultaneously are shown in Figure 3 (Supplementary Data 7D, 8Q). P. × rasumovskoe formed one cluster (I) with the majority of P. nigra samples. It can be assumed that the female parental form for P. × rasumovskoe was P. nigra rather than the second probable ancestor, P. suaveolens (Mayorov et al., 2020). Cluster II included P. × sibirica samples which were mixed with several accessions of P. suaveolens (together with P. koreana and P. maximowiczii), significant number of P. laurifolia, P. longifolia, and P. × wobstii accessions, as well as several accessions of P. nigra and P × irtyschensis. Cluster III with most P. × petrovskoe samples also included a number of samples of P. laurifolia, which along with P. × canadensis is the probable parental species of P. × petrovskoe (Mayorov et al., 2020). Therefore, the probable female parental form for P. × petrovskoe was P. laurifolia rather than another putative ancestor, P. × canadensis. An analysis of the similarity of P. × rasumovskoe and P. × petrovskoe accessions and their probable parental forms based on genetic distances also showed that P. nigra was a more likely female ancestor of P. × rasumovskoe than P. suaveolens (the average genetic distance between accessions of P. × rasumovskoe and P. nigra was 0.7, and that between accessions of P. × rasumovskoe and P. suaveolens was 2.6), and P. laurifolia was a more likely female ancestor of P. × petrovskoe than P. × canadensis or another representative of the section Aigeiros (the average genetic distance between P. × petrovskoe and P. laurifolia samples was 0.5, and that between samples of P. × petrovskoe and P. × canadensis/P. nigra was 2.2) (Supplementary Data 5). Several accessions of P. suaveolens were also included in cluster III. Cluster IV combined subclusters of P. suaveolens (together with P. koreana and P. maximowiczii), P. simonii, and P. talassica, as well as accessions of P. deltoides and its hybrids (including P. × canadensis), P. afghanica and several P. nigra accessions, which were interspersed with each other.
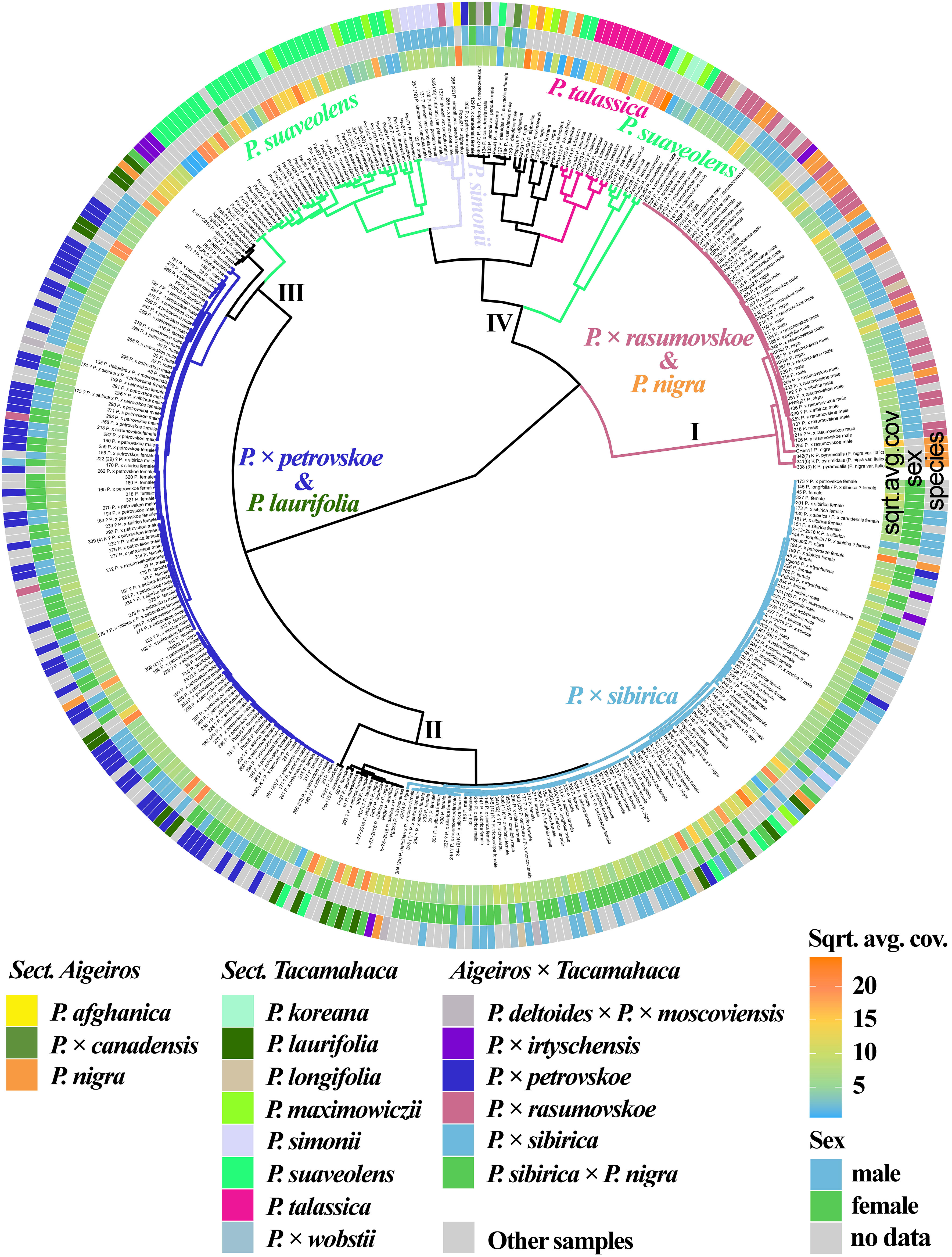
Figure 3 Dendrogram based on deep sequencing data for trnG‐psbK-psbI, rps2-rpoC2, and rpoC2-rpoC1 sequences. Colors corresponding to species and hybrids mark only accessions for which there were no doubts in the morphological determination of the species affiliation.
Thus, the analysis by individual loci used in phylogenetic studies of Populus species, for a number of studied samples of sections Aigeiros and Tacamahaca, corresponded to the views on phylogenetic relationships in the genus Populus, but was significantly less informative than the joint analysis by 14 loci at once.
3.3 Analysis of the SDR and the ARR17 gene
We also analyzed the genetic proximity of poplars based on the sequencing of the loci of the SDR localized on chromosome 19 (Figure 4, Supplementary Data 7E, 8R). It should be explained that, for poplars of sections Aigeiros and Tacamahaca, the part of the SDR comprising partial repeats of the ARR17 gene is present only in male plants, and another part is present in plants of both sexes, but differs significantly in polymorphisms (Zhou et al., 2020). The samples were divided into three large clusters, one of which (I) included only female plants and the other two (II and III) included only male plants (the only exception was sample 355(17) P. × wobstii). A subcluster of male samples of P. × petrovskoe was clearly distinguished in cluster II. A subcluster of a significant number of samples of P. nigra and P. nigra × P. pyramidalis, most male samples of P. × sibirica, as well as P. deltoides and P. × canadensis, was also separated. This subcluster merged with the P. × petrovskoe subcluster into one cluster (II). Based on these data, we may assume that in the male samples of P. × petrovskoe and P. × sibirica, the male parental form was a representative of the section Aigeiros (according to the morphological data, this is most likely P. × canadensis for P. × petrovskoe and P. nigra for P. × sibirica (Mayorov et al., 2020)). The P. × rasumovskoe subcluster was identified in cluster III. Most male poplars of the section Tacamahaca, including P. suaveolens, which is the probable parental form of P. × rasumovskoe, were located in the same cluster with P. × rasumovskoe. Based on the data obtained, we can assume that P. suaveolens was the male parental species of P. × rasumovskoe. Analysis of the similarity of P. × rasumovskoe and P. × petrovskoe samples and their probable parental forms based on genetic distances also showed that P. suaveolens is a more probable male ancestor of P. × rasumovskoe than P. nigra (the average genetic distance between samples of P. × rasumovskoe and P. suaveolens in the male cluster was 1.7, and that between samples of P. × rasumovskoe and P. nigra in the male cluster was 2.3), and P. × canadensis or another representative of the section Aigeiros is a more likely male ancestor of P. × petrovskoe than P. laurifolia (the average genetic distance between P. × petrovskoe and P. × canadensis/P. nigra of the male cluster was 1.8, and that between P. × petrovskoe and P. laurifolia of the male cluster was 2.6) (Supplementary Data 5). Besides, in cluster III of male samples, a subcluster was identified that included a significant group of interspersed P. suaveolens, P. koreana, and P. maximowiczii, indicating their genetic similarity. A subcluster of P. simonii was also revealed in cluster III. Thus, cluster III included male samples of P. × rasumovskoe, P. suaveolens (including P. maximowiczii and P. koreana), P. simonii, P. laurifolia, P. talassica, and P. longifolia which were absent in cluster II of male samples. Within group of male genotypes (clusters II and III), several accessions of P. nigra were revealed in cluster III, but most P. nigra samples were in cluster II.
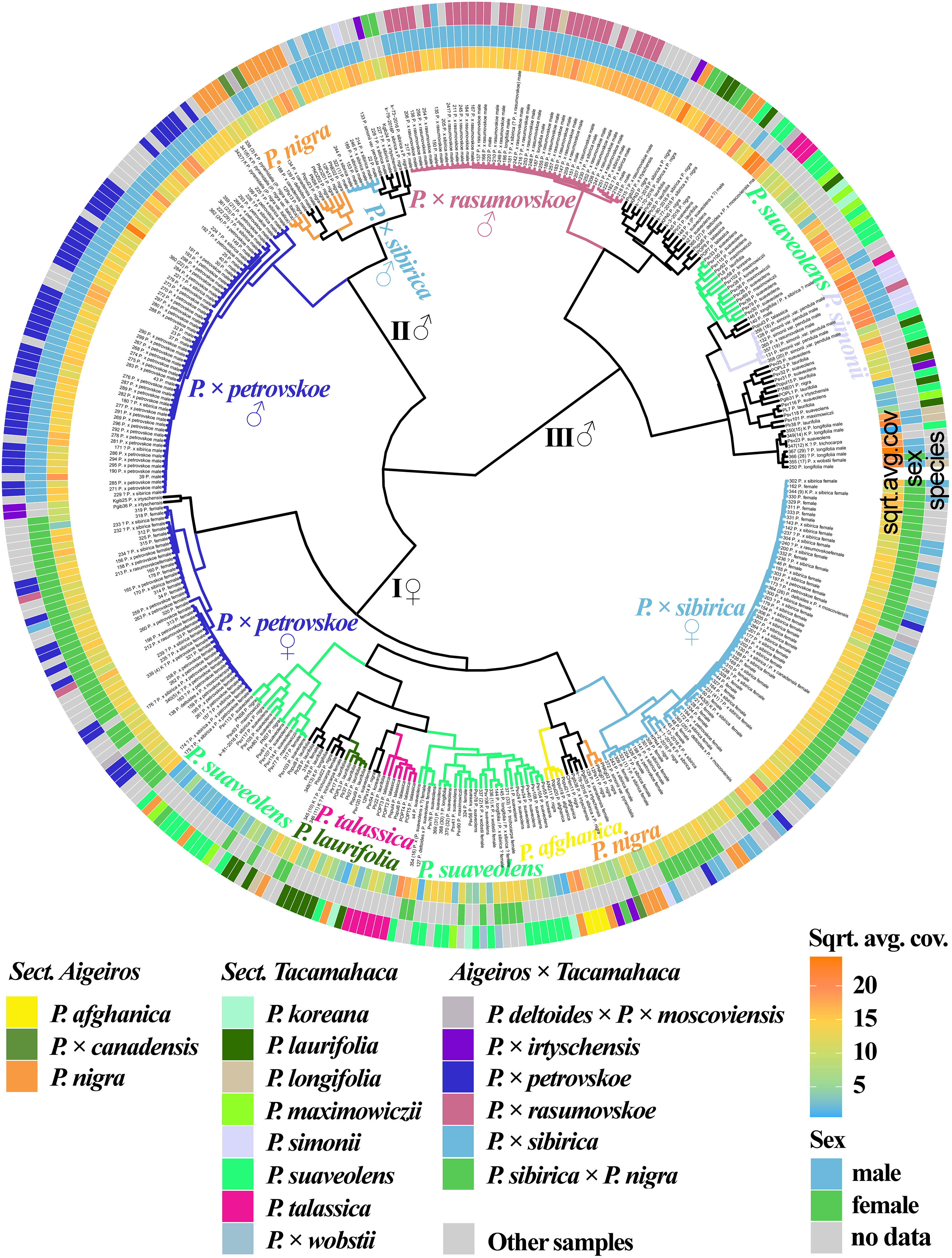
Figure 4 Dendrogram based on deep sequencing data for the SDR sequences. Colors corresponding to species and hybrids mark only accessions for which there were no doubts in the morphological determination of the species affiliation.
In cluster I of female plants, a subcluster of the majority of P. × sibirica samples was separated, which was close to the subcluster of several samples of P. nigra and P. afghanica. A subcluster of female samples of P. × petrovskoe was also distinguished, which, unlike the male samples of this hybrid, was significantly distant from the cluster of P. × sibirica. In addition, the cluster of female accessions included subclusters of accessions of one species, namely P. laurifolia and P. talassica, and two subclusters of mainly P. suaveolens (including P. maximowiczii and P. koreana). However, several samples of P. nigra and P. × wobstii were also in subclusters of P. suaveolens.
The data obtained for the SDR significantly complement the information on the phylogenetic relationships of the studied species and hybrids of sections Aigeiros and Tacamahaca, allowing us to determine the ancestors of hybrids in the male lineage, as well as clearly separate samples into male and female ones.
We also performed clustering based on the data obtained for the ARR17 gene (chromosome 19) (Supplementary Data 7F, 8S). A subcluster of the majority of P. nigra samples was distinguished. A subcluster of most female and male samples of P. × sibirica, which was located in the same cluster as the P. nigra subcluster, was also identified. P. × rasumovskoe samples formed a separate subcluster. P. × petrovskoe divided into two clusters according to sex. A large subcluster consisted mainly of P. longifolia, two distant subclusters of P. suaveolens (including P. maximowiczii and P. koreana), and subclusters of P. talassica and P. simonii were also distinguished. Samples of P. afghanica formed a subcluster close to the P. deltoides and P. × canadensis subcluster, as well as the P. talassica subcluster. The ARR17 analysis allowed us to differentiate samples of many species and hybrids into separate subclusters and was quite informative.
3.4 Joint analysis based on 14 loci traditionally used in poplar phylogenetic studies, SDR, and ARR17 gene
Figure 5 (Supplementary Data 7G, 8T) shows a dendrogram constructed on the basis of all the sequencing data we obtained, namely 14 loci that were previously used in phylogenetic studies of Populus and loci of the SDR and ARR17 gene. Five large clusters were identified. Cluster I included mostly species. Subclusters of cluster I included mainly samples of only one species, namely P. talassica, P. afghanica, P. simonii, and P. longifolia (in this subcluster P. × wobstii and P. trichocarpa was located). Three more subclusters were also distinguished, in each of which samples of P. suaveolens, P. maximowiczii, and P. koreana were mixed. This supports the hypothesis that P. koreana and P. maximowiczii should not be distinguished as species or even subspecies, but should be assigned to P. suaveolens (Skvortsov and Belyanina, 2006). The subcluster of P. afghanica was merged with the subcluster of P. talassica. According to some hypotheses, P. afghanica can be regarded as a hybridogenic species derived from P. nigra and P. talassica (see Supplementary Data 2 for details), and the results of the genetic analysis support the relationship between P. afghanica and P. talassica. Cluster II included samples of P. × rasumovskoe, as well as several samples of P. suaveolens, P. × sibirica × P. nigra, and P. nigra. All samples in this cluster for which the sex was determined were males. Cluster III was represented by predominantly male samples of P. × petrovskoe, which formed a separate subcluster. This cluster also included a significant number of P. nigra accessions, accessions of P. × canadensis (the probable ancestor of P. × petrovskoe), and several male P. × sibirica accessions that formed separate subclusters. In cluster IV, female samples of P. × petrovskoe and most samples of its probable ancestor P. laurifolia formed separate subclusters. Cluster V was represented mainly by female genotypes of P. × sibirica. In the joint analysis, subclusters of individual species and hybrids were distinguished quite well, the picture was clearer than in the analysis of individual loci/groups of loci. This dendrogram most accurately reflected the known data on the phylogenetic relationships of species and hybrids of sections Aigeiros and Tacamahaca and provided an opportunity to test a number of controversial hypotheses about the origin of the studied poplars.
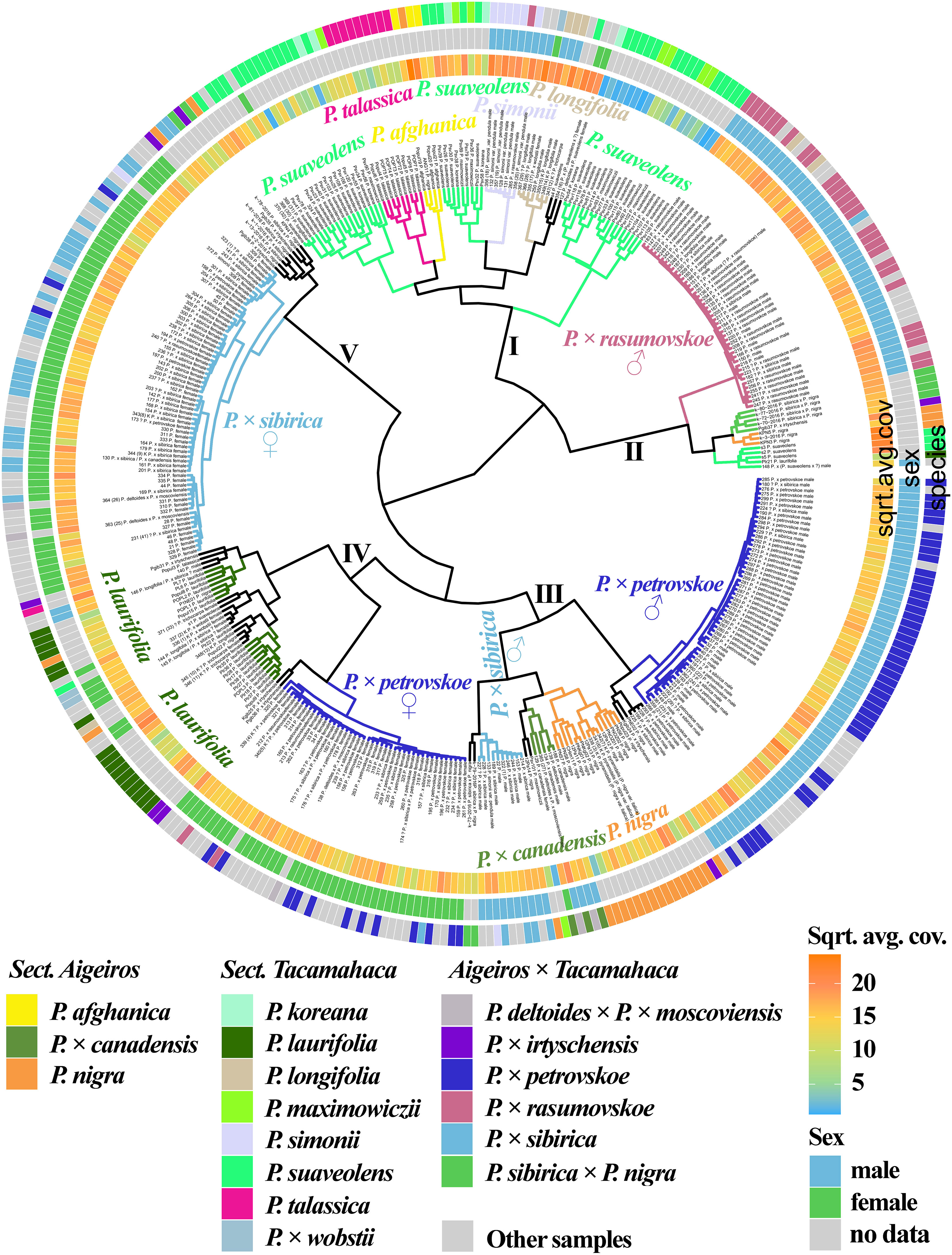
Figure 5 Dendrogram based on deep sequencing data for NTS 5S rDNA, ITS, DSH 2, DSH 5, DSH 8, DSH 12, DSH 29, 6, 15, 16, X18, trnG‐psbK-psbI, rps2-rpoC2, rpoC2-rpoC1, SDR, and ARR17 sequences. Colors corresponding to species and hybrids mark only accessions for which there were no doubts in the morphological determination of the species affiliation.
Thus, the analysis by multicopy and single-copy nuclear DNA sequences, chloroplast DNA sequences, and the SDR loci simultaneously was the most informative and corresponded to the views on the systematics of the studied species and hybrids on the basis of morphological characteristics. Groups of samples of the same species/hybrid were well differentiated into separate subclusters. The results of targeted deep sequencing also shed light on a number of hypotheses derived from the morphological analysis, including the genetic proximity of P. suaveolens, P. koreana, and P. maximowiczii, as well as the relatedness of P. talassica and P. afghanica. The performed genetic analysis made it possible to effectively separate difficult for identification interspecific hybrids P. × rasumovskoe, P. × petrovskoe, and P. × sibirica that grow on the territory of Moscow. In addition, comparison of the results of the analysis by chloroplast DNA and the SDR allowed us to determine which species was the male parental form and which was the female parental form for male samples of P. × rasumovskoe, P. × petrovskoe, and P. × sibirica.
3.5 Clarification of poplar hybrid constitution based on targeted deep sequencing data
In our sample set, accessions of poplar species, especially collected in natural areas, were identified accurately enough on the basis of morphological features. However, for samples collected on the territory of Moscow, there were some difficulties with identification on the basis of morphological characteristics, which were especially evident for accessions of hybrid origin. First of all, this problem is related to cardinal pruning of trees, which is often used in cities, including to control poplar fluff. After such pruning, morphological features typically used for identification of hybrids may be distorted for some time, in particular, shape of crown, presence of shoots and leaves characteristic to a hybrid. As a result, in our sample set there were poplar accessions whose species affiliation could not be established, as well as poplar accessions whose species affiliation was doubted. Taking into account the fact that our genetic analysis allowed us to differentiate well the samples of different species and hybrids, we set the task to determine the species affiliation of those samples for which estimation of species affiliation by morphological features was difficult or was not carried out. Such analysis turned out to be especially relevant for probable hybrids collected mainly in Moscow.
In the dendrogram based on all sequenced loci (Figure 5, Supplementary Data 7G, 8T), the P. × rasumovskoe subcluster contained 13 accessions with unknown or doubtful species affiliation: 150, 151, 181, 182, 215, 216, 217, 218, 219, 220, 223, 230, 241. When analyzed separately by the SDR (Figure 4, Supplementary Data 7E, 8R) and by the set of 14 loci used in phylogenetic studies of poplars (Figure 1, Supplementary Data 6A, 8A), these samples were also in the P. × rasumovskoe cluster, and not in other clusters. It is worth noting that the analysis of the genetic distances between the accessions (Supplementary Data 5) showed that all samples of P. × rasumovskoe were genetically very close, and samples from their cluster with uncertain species affiliation were also close to them. It can be assumed that they belong exactly to P. × rasumovskoe, and accessions of this hybrid distributed in Moscow are apparently descendants of the same clone. In addition, the P. × rasumovskoe group included accessions 186 and 187 classified as P. longifolia according to morphological characteristics. However, according to the SDR loci (Figure 4, Supplementary Data 7E, 8R), the 3 chloroplast genome regions (Figure 3, Supplementary Data 7D, 8Q), the ARR17 gene (Supplementary Data 7F, 8S), and the joint analysis of 14 loci used in poplar phylogenetic studies (Figure 1, Supplementary Data 6A, 8A), these two samples were part of the P. × rasumovskoe cluster. A more detailed analysis of the morphological data on these accessions raised the question of the accuracy of their identification – it is very likely that they morphologically also fit the description of P. × rasumovskoe subjected to cardinal pruning. Accession 205, morphologically identified as P. × sibirica, also fell into the P. × rasumovskoe group rather than into the P. × sibirica group when analyzed on the basis of different sequenced regions. We can assume an error in the identification of this accession on the basis of morphological characteristics due to cardinal pruning as well.
In the dendrogram based on all sequenced loci (Figure 5, Supplementary Data 7G, 8T), the subcluster of male P. × petrovskoe genotypes contained 18 samples with unknown or doubtful species affiliation: 23, 25, 30, 32, 37, 39, 40, 43, 149, 171, 180, 192, 221, 222 (29), 224, 225, 226, 229. The same pattern was observed when analyzed separately for the SDR (Figure 4, Supplementary Data 7E, 8R) and the 14 loci used in poplar phylogenetic studies (Figure 1, Supplementary Data 6A, 8A). It is likely that these 18 accessions belong to P. × petrovskoe, and the male genotypes of P. × petrovskoe are genetically close (Supplementary Data 5) and may be descendants of the same Moscow clone.
In the dendrogram based on all sequenced loci (Figure 5, Supplementary Data 7G, 8T), the subcluster of female genotypes of P. × petrovskoe contained 27 samples with unknown or doubtful species affiliation: 33, 34, 157, 160, 163, 174, 175, 176, 178, 232, 233, 234, 235, 239, 312, 313, 314, 315, 316, 317, 318, 319, 320, 321, 325, 339 (4) K, 340 (5) K. In addition, samples 212 and 213 (identified as P. × rasumovskoe), 138 (P. deltoides × (P. laurifolia × P. suaveolens)), and 170 (P. × sibirica) fell into the same subcluster. In the analysis based on the 14 loci used in phylogenetic studies of poplars (Figure 1, Supplementary Data 6A, 8A), the subcluster of P. × petrovskoe female samples included all the same accessions. When analyzed only by the SDR loci (Figure 4, Supplementary Data 7E, 8R), all samples except 316 and 317 were also included in the P. × petrovskoe female genotype cluster (316 was in the group with P. laurifolia, and 317 was in the group with P. suaveolens). Most likely, 25 of 27 uncertain accessions belong to P. × petrovskoe. Samples 212 and 213, identified as P. × rasumovskoe (the remaining accessions of this hybrid formed a dense subcluster distant from P. × petrovskoe), and sample 170, identified as P. × sibirica (most accessions of this hybrid also formed a detached subcluster), also belong to P. × petrovskoe. For samples 138, 316, and 317, further analysis is required for final conclusions. It should be noted that in the cluster of most female accessions of P. × petrovskoe, there were more difficulties in species identification by morphological features than in the cluster of male P. × petrovskoe genotypes – in the first cluster, a significant number of samples were represented by genotypes with uncertain or doubtful species affiliation, including doubtful P. × sibirica accessions. At the same time, accessions of the subcluster of female P. × petrovskoe genotypes were genetically very close (Supplementary Data 5), and there is a high probability that they are the descendants of the same clone distributed in Moscow.
In the cluster of the majority of female accessions of P. × sibirica when analyzed by all sequenced loci (Figure 5, Supplementary Data 7G, 8T), there were also 33 samples with unknown or doubtful species affiliation: 21, 28, 44, 45, 46, 48, 50, 130, 162, 173, 203, 204, 231, 236, 237, 238, 240, 264, 308, 309, 310, 311, 323 (1), 326, 327, 328, 329, 330, 331, 332, 333, 334, 335. In addition, this cluster included accessions of P. deltoides × P. × moscoviensis (363 (25) and 364 (26)), P. × petrovskoe (194, 197, 198), and P. simonii var. fastigiata (372). When analyzed only by the SDR (Figure 4, Supplementary Data 7E, 8R), the same samples were also in the cluster of female P. × sibirica accessions. When analyzed on the basis of the 14 loci used in poplar phylogenetic studies (Figure 1, Supplementary Data 6A, 8A), 30 of the 33 poplars with uncertain species affiliation (except for 21, 328, 329) were also in the P. × sibirica cluster. It is likely that these 30 accessions belong to P. × sibirica. In addition, errors in the identification of samples 194, 197, 198 as P. × petrovskoe (since most female accessions of this hybrid formed a separate cluster, and after cardinal pruning this hybrid could be confused with P. × sibirica) and error at the stage of sample preparation for accession 372, listed as P. simonii var. fastigiata (this species is difficult to confuse with P. × sibirica by its morphological features). For accessions of P. deltoides × P. × moscoviensis (363 (25) and 364 (26)), the question of the accuracy of identification remains open. For female accessions of P. × sibirica, we observed a different pattern from P. × rasumovskoe and P. × petrovskoe: for this group, the samples were not as genetically close to each other as in P. × rasumovskoe and P. × petrovskoe cases, and they probably originated not from one but from several original clones.
It is worth noting that in the analysis based on all loci we sequenced (Figure 5, Supplementary Data 7G, 8T), a subcluster close to the main subcluster of P. × sibirica female samples was formed by P. × irtyschensis (Pgib35 and Pgib38), P. × sibirica × P. nigra (k-78-2016), P. nigra (KPN4 and k-2-2-2016), and P. × sibirica (k-1-2016 and k-13-2016). When analyzing only the SDR loci (Figure 4, Supplementary Data 7E, 8R), P. nigra (KPN4 and k-2-2-2016) formed a separate subcluster with P. × sibirica (k-1-2016 and k-13-2016) accessions in a large subcluster of P. × sibirica, but P. × irtyschensis (Pgib35 and Pgib38) and P. × sibirica × P. nigra (k-78-2016) samples were in a different subcluster and were closer to P. afghanica than to the main P. × sibirica subcluster. When the 14 loci used in poplar phylogenetic studies were analyzed (Figure 1, Supplementary Data 6A, 8A), the picture for the group of female P. × sibirica accessions was slightly different from those obtained on the basis of the SDR or all sequenced loci. P. × sibirica (k-1-2016 and k-13-2016) and P. nigra (KPN4 and k-2-2-2016) samples moved to a large subcluster of P. × sibirica × P. nigra and P. × irtyschensis samples. P. × sibirica samples k-1-2016 and k-13-2016 were collected in the Novokuznetsk district of Kemerovo Region, in contrast to the rest of the P. × sibirica samples collected in Moscow. This may explain their clustering with P. × sibirica × P. nigra and P. × irtyschensis, also collected in the Novokuznetsk district. Thus, the relationship between the geographical origin of the samples and genotype was revealed for these accessions.
In our sample set we had two accessions from Uzbekistan (ANG11 and CHim11), which were listed in the herbarium as P. nigra, but could belong to P. afghanica, because they were collected outside the area of P. nigra, and these two species are not always distinguishable by a single shoot on a herbarium leaf. ANG11 was in the same cluster as the three P. afghanica accessions we studied, and probably belongs to this species rather than to P. nigra. CHim11 clustered with the rest of the P. nigra samples and indeed belongs to this species.
In addition, a clear error, probably associated with sample preparation, is related to sample 265 (possibly confused with sample 372), identified as P. × rasumovskoe, which was in the subcluster of P. simonii accessions in the analysis for both all loci studied and individual loci. P. × rasumovskoe and P. simonii are morphologically very different, so an error in the morphological determination of species affiliation is unlikely.
Thus, we have shown a high informativity of the use of the sex-determining region to study the relationships between representatives of sections Aigeiros and Tacamahaca, especially interspecific hybrids, which are difficult for systematics. Joint analysis of the targeted deep sequencing data for the SDR and ARR17 gene, as well as for the used in molecular phylogeny sequences of NTS 5S rDNA, ITS, DSH 2, DSH 5, DSH 8, DSH 12, DSH 29, 6, 15, 16, X18, trnG-psbK-psbI, rps2-rpoC2, rpoC2-rpoC1 was informative in determining the taxonomic affiliation of poplar accessions.
4 Discussion
To study the genetic relationship of poplar species and hybrids from sections Aigeiros and Tacamahaca, we chose an approach based on targeted deep sequencing of amplicons. This method drastically reduces the cost of analysis per sample compared to whole genome sequencing, has incomparable accuracy in the study of heterozygous genotypes and multicopy sequences, as well as less laboriousness in the analysis of large sample sets for a significant number of loci compared to Sanger sequencing (McCormack et al., 2013; Cruaud et al., 2017; Borkhert et al., 2018; Melnikova et al., 2019; Dmitriev et al., 2020; Novakovskiy et al., 2020; Povkhova et al., 2022). However, effective application of targeted deep sequencing requires the selection of the most informative loci.
Compared to the previous studies on Populus species based on the analysis of multicopy and single-copy sequences of nuclear and chloroplast genomes (Schmidt et al., 1994; Leskinen and Alström-Rapaport, 1999; Page, 2000; Huson and Scornavacca, 2011; Schroeder et al., 2012; Huang et al., 2014; Wang et al., 2014; Alexandrov and Karlov, 2018; Borkhert et al., 2018; Zhou et al., 2018; Wang et al., 2019), we sequenced regions of the SDR and the ARR17 gene, which is the key gene in sex determination in the genus Populus (Muller et al., 2020). The SDR allowed us to reveal the origin of the examined genotypes in the male lineage and significantly complemented the results obtained in the analysis of the DNA sequences traditionally used in poplar studies.
We studied interspecific hybrids, including those created by breeders, actively used in landscaping, as well as hybrids of natural origin and “pure” species from sections Aigeiros and Tacamahaca, which have an XY sex-determination system and interbreed easily. In poplars of these sections, the SDR is localized on the peritelomeric region of chromosome 19 and includes male-specific region containing partial repeats of the ARR17 gene, and a region that is present in plants of both sexes but differs significantly in DNA polymorphisms between males and females (Muller et al., 2020; Xue et al., 2020; Zhou et al., 2020; Melnikova et al., 2021; Pushkova et al., 2021), which is probably associated with the suppression of recombination. In this regard, the SDR could be promising for use in phylogenetic studies of species and hybrids of sections Aigeiros and Tacamahaca. Previously, based on whole-genome sequencing data, we showed the informativity of the SDR polymorphisms for determining putative parental forms of poplar hybrids (Melnikova et al., 2021). However, this approach requires significant costs for the analysis of one sample, so the analysis of large sample sets is complicated. Therefore, in this work, we designed primers for the amplification of the SDR loci of poplars of sections Aigeiros and Tacamahaca containing sex-specific polymorphisms (Supplementary Data 3) for subsequent targeted deep sequencing.
Analysis of the SDR sequences made it possible to divide the studied samples of sections Aigeiros and Tacamahaca into male and female, and within groups of the same sex, the samples were predominantly clustered according to belonging to a particular hybrid or species (Figure 4, Supplementary Data 7E, 8R). In addition to the SDR, we also sequenced DNA sequences traditionally used in poplar phylogenetic studies, namely NTS 5S rDNA, ITS, DSH 2, DSH 5, DSH 8, DSH 29, 6, 15, 16, X18, trnG-psbK-psbI, rps2-rpoC2, rpoC2-rpoC1 (Schmidt et al., 1994; Schroeder et al., 2012; Huang et al., 2014; Wang et al., 2014; Zhou et al., 2018; Wang et al., 2019). Clustering based on multicopy (NTS 5S rDNA and ITS) and single-copy (DSH 2, DSH 8, DSH 29, 6, 15, 16, X18) sequences of the nuclear genome and regions of the chloroplast genome (trnG-psbK-psbI and rps2-rpoC2) made it possible to separate a number of samples into groups according to belonging to a particular hybrid (often) or species (less often), however, none of the sequences allowed us to obtain a clear picture corresponding to the understanding of the similarity of the studied accessions based on morphological data. The most complete information on the phylogenetic relationships of the studied poplar accessions was achieved through the joint analysis of data for multicopy and single-copy sequences of nuclear DNA, loci of chloroplast DNA, and the SDR – clustering based on all obtained data of targeted deep sequencing most closely corresponded to the ideas about systematics of poplars based on morphological characteristics (Figure 5, Supplementary Data 7G, 8T). Since the results of genetic analysis corresponded to the known facts, we can assume that they can be used to test hypotheses about the origin of poplars, as well as to determine the species identity of accessions whose identification on the basis of morphological features was complicated.
The analysis of sequencing data for the SDR and chloroplast genome loci made it possible to determine the origin of P. × petrovskoe [P. laurifolia (female tree) × P. × canadensis (male tree)] and P. × rasumovskoe (P. nigra (female tree) × P. suaveolens (male tree). The original description of P. × rasumovskoe given by M. Wolkenstein is as follows: “A hybrid between P. nigra, fertilised with the pollen of P. suaveolens” (Wolkenstein, 1882). It agrees with the results obtained by us on the basis of genetic analysis. However, for P. × petrovskoe, the original description by M. Wolkenstein is as follows: “A hybrid between P. canadensis fertilised by the pollen of P. suaveolens” (Wolkenstein, 1882). The indication of P. suaveolens as the parental species for P. × petrovskoe is a probable mistake by M. Wolkenstein. The Russian poplar species of the section Tacamahaca, of which there are two, were primarily involved in hybridization in Russia, and the “balsamic” component in P. × petrovskoe is probably represented by P. laurifolia, which is also proved morphologically by the ribbing of shoot axes (Mayorov et al., 2020). The sequencing data also confirm the error in the original description of P. × petrovskoe. The first parental species, P. × canadensis, was indicated correctly by Wolkenstein, although it went to the hybrid in the male lineage.
We analyzed the relationships of the studied poplar species and hybrids of sections Aigeiros and Tacamahaca based on molecular genetic (obtained results of targeted deep sequencing of NTS 5S rDNA, ITS, DSH 2, DSH 5, DSH 8, DSH 29, 6, 15, 16, X18, trnG-psbK-psbI, rps2-rpoC2, rpoC2-rpoC1, ARR17, and SDR – Figure 5), morphological, and ecological-geographical data. Below is our interpretation of the results obtained for particular poplar species and hybrids.
It is known that not universally recognized P. koreana and P. maximowiczii species are morphologically very close to P. suaveolens or even coincide with it (Skvortsov and Belyanina, 2006). In Figure 5 and dendrograms based on individual loci (Supplementary Data 6, 7), P. koreana and P. maximowiczii accessions always appear together with P. suaveolens ones. Thus, the obtained results indicate not only genetic proximity but also the identity of P. suaveolens, P. koreana, and P. maximowiczii, confirming the controversial assumption of Skvortsov and Belyanina (Skvortsov and Belyanina, 2006), which is still not recognized by most botanists, that P. koreana and P. maximowiczii should not be distinguished as species, but should be assigned to P. suaveolens. If these taxa are understood separately, we can speak only about geographical races, which are strictly confined to certain regions (P. koreana to Korea, P. maximowiczii to the Far East, etc.). It is unfounded to consider, as it is usually done, that all three species grow simultaneously, for example, in the Far East south of Amur. In this case, we can speak only about the external deviation to the appearance peculiar to a certain race.
All P. × canadensis accessions were arranged compactly in Figure 5, with only P. deltoides and its hybrid with P. × moscoviensis falling into their subcluster, and this is natural, since P. × canadensis, according to the well-established notions, is a hybrid of P. deltoides and P. nigra. The distinction between P. × canadensis and P. deltoides is sometimes difficult; in the past, they were generally mixed (Skvortsov, 2010), and even errors in identification are possible. In the neighboring subclusters, exclusively P. nigra and P. pyramidalis appeared, and in the next ones by distance, almost exclusively P. × petrovskoe was observed. P. × petrovskoe, according to our recent assumption (Mayorov et al., 2020), is a hybrid of P. × canadensis and P. laurifolia, and the obtained molecular genetic results confirm this hypothesis. This result is not trivial at all, because previously the putative parental species of P. × petrovskoe were indicated as P. deltoides s.l. × P. suaveolens (Regel, 1889; Wolkenstein, 1882); P. deltoides × P. jackii (Dippel, 1892); P. × canadensis [or also P. deltoides?] × P. suaveolens (Bogdanov, 1965); P. deltoides × P. laurifolia (Koltzenburg, 1999; Tsvelev, 2001). Only Skvortsov (Skvortsov, 2010) indicated the same P. × canadensis and P. laurifolia, but in relation to P. × berolinensis K.Koch, for which this researcher mistakenly took P. × petrovskoe.
Most P. nigra accessions in Figure 5 were compactly arranged in cluster III together with P. pyramidalis. This group is adjacent to P. × canadensis, P. deltoides, and P. × petrovskoe. Several more P. nigra accessions were in other clusters, and for 5 of them, P. × irtyschensis accessions were the closest neighbors. Another close neighbor of P. nigra is P. laurifolia, which, according to our conceptions (Nasimovich et al., 2019), is a hybridogenic species that originated with the involvement of P. nigra. These results are quite natural, they support theoretical insights, but even more so, they support the ability of the analysis based on the targeted deep sequencing of the SDR and other nuclear and chloroplast loci used in phylogenetic studies to examine poplars. Importantly, samples of P. nigra and closely related species (primarily P. × irtyschensis) formed joint compact groups simultaneously in three different clusters (II, III, V). This suggests that the distribution of accessions, in addition to species affiliation, is influenced by the general polymorphism of P. nigra [also shown by Wang et al. (Wang et al., 2022)] and closely related species, and this must be taken into account when working with poplars. It is also important that accessions of P. nigra and closely related species did not fall into cluster I, formed primarily by P. suaveolens (including P. koreana and P. maximowiczii). This confirms the idea expressed earlier (Nasimovich and Vasilyeva, 2019), that P. nigra and P. suaveolens are geographical, biotopic, and morphological antipodes, occupying as if opposite “poles” in the unified system of Eurasian poplars. It turns out that they are also genetic “poles” of this system.
All but one P. laurifolia accessions in Figure 5 formed a compact group of subclusters in cluster IV, isolated from P. nigra and closely related species, and isolated from P. suaveolens and closely related species (cluster I). This probably implies the species independence of P. laurifolia, although we have argued that P. laurifolia emerged as a hybridogenic species at the intersection of gene flow from P. nigra and P. suaveolens (Nasimovich et al., 2019). Based on the molecular genetic analysis, P. laurifolia is slightly closer to P. nigra than to P. suaveolens. P. laurifolia is one of the “balsam” poplars, but partially shares a common habitat with P. nigra, which turns out to be more important. In the same or close subclusters, together with P. laurifolia, there were its putative hybrids: P. × irtyschensis (P. laurifolia × P. nigra – 1 sample) and P. × wobstii (P. laurifolia × P. longifolia – 2 samples), which confirms their hybrid nature and the putative composition of parental species. For P. × wobstii, this result is not quite trivial, because previously the putative parental species of P. × wobstii were indicated as P. suaveolens (Regel, 1889); P. balsamifera (Dippel, 1892); P. suaveolens and P. jackii (Sokolov et al., 1951); P. simonii and P. suaveolens (Karhu and Hamet-Ahti, 1992); P. laurifolia and P. tristis (Rehder, 1949; Koltzenburg, 1999); P. laurifolia and P. tristis (or P. longifolia) (Ascherson and Graebner, 1908-1913; Tsvelev, 2001).
Most P. longifolia accessions in Figure 5 were in cluster I together with P. suaveolens and closely related species, but they were absent in clusters III (together with P. nigra) and IV (together with P. laurifolia). This indirectly confirms our hypothesis on the origin of P. longifolia from P. suaveolens (Mayorov et al., 2012), although for complete certainty it is necessary to investigate in the same way the similarity with the American species P. balsamifera and P. trichocarpa, which are also considered as ancestors of P. longifolia (Tsinovskis, 1977; Skvortsov, 2008).
P. suaveolens and closely related species constitute the vast majority in cluster I (Figure 5) and, in addition, are present in cluster II (3 accessions) together with P. × rasumovskoe, while in clusters III and IV, where P. nigra and P. laurifolia dominate among “pure” species, they are absent. This means a molecular genetic opposition with respect to other (more western) “pure” species, as already mentioned above. P. longifolia, P. talassica, P. afghanica, and the hybrid P. × rasumovskoe (P. nigra × P. suaveolens), which form clusters I and II, are also close to P. suaveolens (together with P. koreana and P. maximowiczii). P. talassica has even been combined with P. suaveolens into one species in the past; P. afghanica is geographically close to P. suaveolens; P. longifolia may have separated from P. suaveolens; and the situation with P. × rasumovskoe is even clearer (see above). In general, the position of P. suaveolens on the dendrogram is completely logical and shows great opportunities for the use of targeted deep sequencing of chloroplast and nuclear sequences, including the SDR, in the study of poplars. Analysis of sequencing data revealed the genetic proximity of P. talassica and P. afghanica. These results support the hypothesis of hybridogenic origin of P. afghanica – this species is close to P. nigra by the traits of leaves and shoots, but there are also traits characteristic of poplars of the section Tacamahaca, probably derived from P. talassica (Supplementary Data 2). It can also be seen that the geographical proximity of species and, therefore, the possibility of hybridization between them is sometimes more important in terms of genetics than the sectional position of species: P. suaveolens and P. afghanica belong to different sections, but are geographically close and therefore alike.
Accessions of the complex hybrid P. × petrovskoe, together with its putative parental species, P. deltoides and P. nigra, form the core of neighboring clusters III and IV (Figure 5), which proves their kinship. Another putative parental species of P. × petrovskoe, P. laurifolia, is also a part of cluster IV. It is also important that P. × petrovskoe does not overlap with P. suaveolens (clusters I and II) in Figure 5, and it is the only mass hybrid of Moscow landscaping whose parental species, according to our knowledge (Mayorov et al., 2020), does not include P. suaveolens. Thus, the molecular genetic analysis fully confirmed our ideas about P. × petrovskoe.
Based on the results of targeted deep sequencing and on the analysis of morphological and ecological-geographical data (see Supplementary Data 2), we present a scheme of relationships between species and hybrids of sections Aigeiros and Tacamahaca (Figure 6). It should be noted that relationships between taxa from different sections are traced. P. afghanica (Aigeiros) is mountain poplar and morphologically close to P. nigra (Aigeiros), but also has some features similar with “balsam” poplars. We supposed that P. afghanica can be a species of hybrid origin that arose at the intersection of gene flow from P. nigra and P. talassica (Tacamahaca). The obtained genetic data support this hypothesis. P. laurifolia (Tacamahaca), which is a fairly typical representative of “balsam” poplars and has all diagnostic features of this section, is genetically close to P. nigra (Aigeiros), but distant from another “balsam” poplar, P. suaveolens (Tacamahaca). In addition, a significant number of poplar hybrids, including P. × petrovskoe, P. × rasumovskoe, P. × sibirica, and P. × irtyschensis, come from crosses of representatives of different sections.
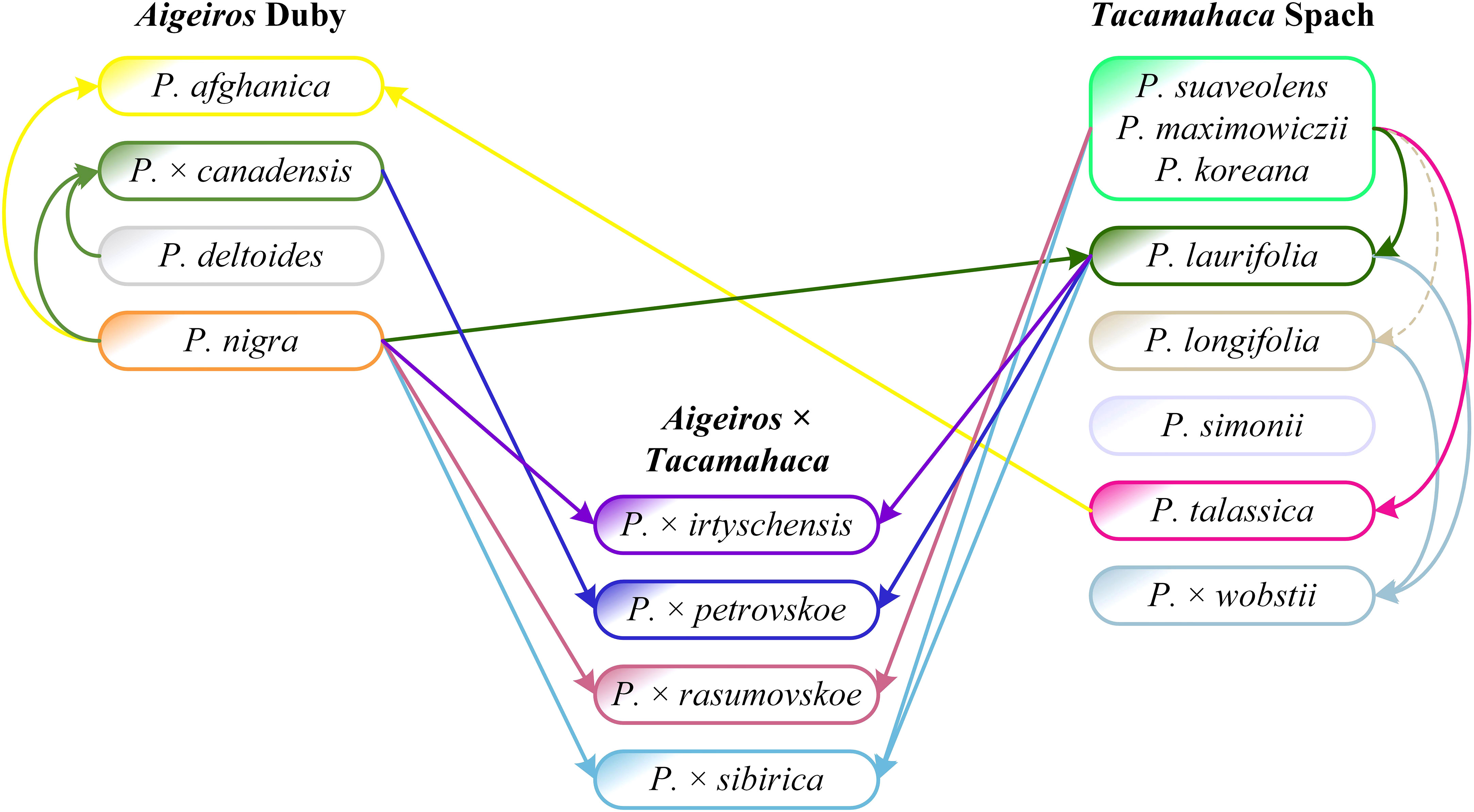
Figure 6 Scheme of relationships between species and hybrids of sections Aigeiros and Tacamahaca. Arrows connect a probable ancestor and a descendant, are directed from an ancestor to descendant, and have the same color as a probable descendant. A dotted arrow indicates that there are doubts about the relationship.
In the group of “black” poplars (Aigeiros), there are similarities of P. × canadensis with P. nigra and P. deltoides. The genetic data are consistent with the known information that P. nigra and P. deltoides are the parental species of P. × canadensis. In the group of “balsam” poplars (Tacamahaca), it is inexpedient to distinguish P. suaveolens, P. koreana, and P. maximowiczii as separate species – they are a single species according to both morphological (Skvortsov and Belyanina, 2006) and genetic data. P. suaveolens is a prominent representative of “balsam” poplars growing in mountain conditions. It is probably the parental species for P. laurifolia, P. talassica, and possibly P. longifolia based on the analysis of both morphological and genetic data. P. × wobstii probably comes from crosses between P. laurifolia and P. longifolia. According to genetic data, P. simonii is close to P. suaveolens, which can be explained by the ecological-geographical relationship of these species.
Considering the status of sections Aigeiros and Tacamahaca, it should be noted that their representatives freely interbreed in both cultivation and nature. Meanwhile, sections Aigeiros and Tacamahaca differ in a large set of traits (more details in Materials and Methods), but all these traits are adaptations to either plain (“black” poplars) or mountain conditions (“balsam” poplars) (Nasimovich et al., 2019). The traits of “black” poplars are most pronounced in P. nigra growing on the vast Russian plain, while the traits of “balsam” poplars are most expressed in P. suaveolens growing in the harsh mountain conditions of the Eastern Siberia (Nasimovich and Vasilyeva, 2019). Our study revealed significant genetic differences between P. nigra and P. suaveolens. Considering a number of other poplar species (e.g., P. laurifolia and P. afghanica), the genetic unity within sections is not revealed. Geographical proximity and the possibility of gene exchange turn out to be more important in terms of genetics than the sectional position. Therefore, sections Aigeiros and Tacamahaca are most likely ecological (plain and mountain poplars) and have no phylogenetic or genetic meaning. However, discarding these sections would make the taxonomy of poplars less convenient, as we discussed earlier (Nasimovich et al., 2019).
The performed genetic analysis also made it possible to establish belonging to the common Moscow hybrids, P. × rasumovskoe, P. × petrovskoe, and P. × sibirica, for a number of accessions with unknown or doubtful origins, as well as to find several errors in identification of accessions. To control fluff in Russian cities, poplars are often subjected to cardinal pruning leading to changes in the traits of leaves, shoots, and crowns, which makes it difficult to identify poplar species and hybrids on the basis of morphological characteristics, and for such accessions, genetic analysis can clarify the situation.
Our study allowed us to evaluate the polymorphism of the three hybrids most commonly used in Moscow landscaping: P. × rasumovskoe, P. × petrovskoe, and P. × sibirica. The analysis based on the data for all sequenced regions at once provided the clearest picture of the separation of samples of these hybrids into their respective groups, but even analyses by individual loci in most cases allowed us to separate the P. × rasumovskoe, P. × petrovskoe, and P. × sibirica groups (Figures 1-5, Supplementary Data 6, 7).
P. × rasumovskoe was represented by male accessions that were genetically very close – the average genetic distance between accessions was 0.2 (Supplementary Data 5). P. × rasumovskoe is one of the spontaneous hybrids discovered before 1882 by R.I. Schroder at the Moscow Agricultural Academy (Wolkenstein, 1882). Probably, all accessions of P. × rasumovskoe studied by us are descendants of one clone, which was quite reasonably used in landscaping of Moscow – male plants do not form fluff and have an attractive weeping crown shape.
The studied samples of P. × petrovskoe were both male and female. Most male accessions were genetically close (the average genetic distance between the accessions was 0.2) and were probably the descendants of one clone. However, two other small groups of P. × petrovskoe male accessions were genetically different from the main group (the average genetic distance between them and the main group was 0.8) (Supplementary Data 5). The main group of female trees of P. × petrovskoe also probably came from one clone (the average genetic distance between the accessions was 0.2), and the three genetically different female accessions most likely belong not to P. × petrovskoe but to P. × sibirica. Thus, two clones of P. × petrovskoe were predominantly used in Moscow landscaping – male one and female one. While the use of the male clone is justified, the planting of female plants was a mistake that led to the appearance of poplar fluff in the city during the ripening of seeds (poplar fluff contains seeds).
For P. × sibirica, the picture was different from that obtained for P. × rasumovskoe and P. × petrovskoe. P. × sibirica was represented predominantly by female trees, and not as genetically similar as in the case of P. × rasumovskoe and P. × petrovskoe. It can be assumed that several predominantly female P. × sibirica clones were used in the landscaping of Moscow. Such a choice was unreasonable and led to a massive problem with poplar fluff.
It is worth noting that among Moscow citizens an opinion about the sex change of poplars is widespread, including due to the media. There is a belief that originally only male plants were planted, and then under the influence of adverse factors they have changed sex to a female, and this has led to the problem of poplar fluff, which is formed only on female plants. Our work shows that the cause of the mass appearance of fluff is errors in the choice of initial material for landscaping, and not at all the sex change of poplars. Only in the case of P. × rasumovskoe, descendants of a male clone were used. For P. × petrovskoe, both male and female descendants were planted. And for P. × sibirica, descendants of several female clones were mostly used in landscaping. We observed complete correspondence between the presence of female or male generative organs on P. × rasumovskoe, P. × petrovskoe, and P. × sibirica trees and the division of all samples of these hybrids into two sexes based on the sequences of the SDR. In this regard, the assumption of mass sex change in Moscow poplars is erroneous. The possibility of formation of male, female, and hermaphroditic generative organs on one tree and the change of their ratio during tree development was shown for hybrids of white poplar (Populus alba L.) and aspen (Populus tremula L.) – gray poplar (Populus × canescens (Aiton) Sm.) (Sabatti et al., 2020). This is due to the fact that P. alba has a ZW system of sex determination and P. tremula has an XY system of sex determination (Cronk and Muller, 2020; Muller et al., 2020). P. × canescens, as well as P. alba and P. tremula, occurs in Moscow an order of magnitude less frequently than intersectional hybrids of poplars of sections Aigeiros and Tacamahaca (Nasimovich et al., 2019; Mayorov et al., 2020); they can also be identified using genetic markers (Kim et al., 2021).
Thus, the use of the SDR loci considerably complements the sequences traditionally used for phylogenetic studies to clarify poplar systematics, makes it possible to divide accessions into male and female and determine the ancestral species in the male lineage. We shed light on a number of controversial hypotheses on the division of poplar accessions into species and on the origin of a number of species and hybrids. A detailed analysis of the obtained data on the SDR sequences and the regions of nuclear and chloroplast genomes traditionally used in phylogenetic studies of poplars with involvement of morphological data will make it possible to clarify the origin and systematics of other studied species of the genus Populus as well.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, BioProject PRJNA943410.
Author contributions
AD and NM conceived and designed the work. EB, EP, YN, MK, NV, RM, RN, LP, DZ, AT, ES, AS, and NB performed the experiments. EB, EP, YN, MK, RM, ED, AK, NB, GK, AD, and NM analyzed the data. EB, EP, YN, DZ, GK, AD, and NM wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work was financially supported by the Russian Science Foundation, grant number 22-14-00404.
Acknowledgments
We are grateful to Dr. I.A. Schanzer for substantial consultative assistance in the preparation of the manuscript. We thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, EIMB RAS for providing the computing power and techniques for the data analysis. This work was performed using the equipment of EIMB RAS “Genome” center (http://www.eimb.ru/ru1/ckp/ccu genome ce.php).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1204899/full#supplementary-material
References
Alexandrov, O. S., Karlov, G. I. (2018). Development of 5S rDNA-based molecular markers for the identification of Populus deltoides Bartr. ex Marshall, Populus nigra L., and their hybrids. Forests 9 (10), 604. doi: 10.3390/f9100604
Ascherson, P., Graebner, P. (1908-1913). Synopsis der mitteleuropaischen flora (Leipzig: W. Engelmann).
Bakulin, V. T. (2005). Use of Poplar in the Landscaping of Industrial Cities of Siberia: a Brief Analysis of the Problem [Ispol'zovanie topolya v ozelenenii promyshlennyh gorodov Sibiri: kratkij analiz problem]. Contemp. Problems Ecol. 12 (4), 563–571.
Bogdanov, P. L. (1965). Poplars and their cultivation [Topolya i ih kul'tura] (Moscow: Forest industry).
Borkhert, E., Krasnov, G., Bolsheva, N., Kezimana, P., Yurkevich, O., Muravenko, O., et al. (2018). Genetics polymorphism of poplars from Moscow Region based on high-throughput sequencing of ITS. Vavilov J. Genet. Breed. 22, 531–535. doi: 10.18699/VJ18.391
Cronk, Q. C. (2005). Plant eco-devo: the potential of poplar as a model organism. New Phytol. 166 (1), 39–48. doi: 10.1111/j.1469-8137.2005.01369.x
Cronk, Q., Muller, N. A. (2020). Default sex and single gene sex determination in dioecious plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01162
Cruaud, P., Rasplus, J. Y., Rodriguez, L. J., Cruaud, A. (2017). High-throughput sequencing of multiple amplicons for barcoding and integrative taxonomy. Sci. Rep. 7, 41948. doi: 10.1038/srep41948
DiFazio, S. P., Slavov, G. T., Joshi, C. P. (2011). “Populus: a premier pioneer system for plant genomics,” in Genetics, genomics and breeding of poplar. Eds. Joshi, C., Difazio, S. P., Kole, C. (Lebanon: Science Publishers, Inc), 1–28.
Dmitriev, A. A., Kezimana, P., Rozhmina, T. A., Zhuchenko, A. A., Povkhova, L. V., Pushkova, E. N., et al. (2020). Genetic diversity of SAD and FAD genes responsible for the fatty acid composition in flax cultivars and lines. BMC Plant Biol. 20 (Suppl 1), 301. doi: 10.1186/s12870-020-02499-w
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bull. 19 (1), 11–15.
Du, S., Wang, Z., Zhang, J. (2014). A novel set of single-copy nuclear DNA markers for the genetic study of Salicaceae. Genet. Mol. Res. 13 (3), 4911–4917. doi: 10.4238/2014.July.4.5
Eckenwalder, J. E. (1996). Systematics and evolution of Populus," in Biology of Populus and its implications for management and conservation. Eds. Stettler, R. F., Bradshaw, H. D., Heilman, P. E., Hinckley, T. M. (Montreal, Canada: NRC research Press), 7–32.
Falistocco, E., Passeri, V., Marconi, G. (2007). Investigations of 5S rDNA of Vitis vinifera L.: sequence analysis and physical mapping. Genome 50 (10), 927–938. doi: 10.1139/g07-070
Garrison, E., Marth, G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907v2. doi: 10.48550/arXiv.1207.3907
Gaudet, M., Jorge, V., Paolucci, I., Beritognolo, I., Mugnozza, G. S., Sabatti, M. (2008). Genetic linkage maps of Populus nigra L. including AFLPs, SSRs, SNPs, and sex trait. Tree Genet. Genomes 4 (1), 25–36. doi: 10.1007/s11295-007-0085-1
Geraldes, A., Hefer, C. A., Capron, A., Kolosova, N., Martinez-Nunez, F., Soolanayakanahally, R. Y., et al. (2015). Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol. Ecol. 24 (13), 3243–3256. doi: 10.1111/mec.13126
Hamzeh, M., Dayanandan, S. (2004). Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast TRNT-TRNF region and nuclear rDNA. Am. J. Bot. 91 (9), 1398–1408. doi: 10.3732/ajb.91.9.1398
Hofmeister, B. T., Denkena, J., Colome-Tatche, M., Shahryary, Y., Hazarika, R., Grimwood, J., et al. (2020). A genome assembly and the somatic genetic and epigenetic mutation rate in a wild long-lived perennial Populus trichocarpa. Genome Biol. 21 (1), 259. doi: 10.1186/s13059-020-02162-5
Hsiao, C., Chatterton, N. J., Asay, K. H., Jensen, K. B. (1994). Phylogenetic relationships of 10 grass species: an assessment of phylogenetic utility of the internal transcribed spacer region in nuclear ribosomal DNA in monocots. Genome 37 (1), 112–120. doi: 10.1139/g94-014
Hsiao, C., Chatterton, N. J., Asay, K. H., Jensen, K. B. (1995). Phylogenetic relationships of the monogenomic species of the wheat tribe, Triticeae (Poaceae), inferred from nuclear rDNA (internal transcribed spacer) sequences. Genome 38 (2), 211–223. doi: 10.1139/g95-026
Huang, D. I., Hefer, C. A., Kolosova, N., Douglas, C. J., Cronk, Q. C. B. (2014). Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae). New Phytol. 204 (3), 693–703. doi: 10.1111/nph.12956
Huang, X. Y., Shang, J., Zhong, Y. H., Li, D. L., Song, L. J., Wang, J. (2022). Disaggregation of ploidy, gender, and genotype effects on wood and fiber traits in a diploid and triploid hybrid poplar family. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.866296
Huson, D. H., Scornavacca, C. (2011). A survey of combinatorial methods for phylogenetic networks. Genome Biol. Evol. 3, 23–35. doi: 10.1093/gbe/evq077
Janković, B., Manic, N., Dodevski, V. (2021). Pyrolysis kinetics of Poplar fluff bio-char produced at high carbonization temperature: A mechanistic study and isothermal life-time prediction. Fuel 296, 120637. doi: 10.1016/j.fuel.2021.120637
Jansson, S., Bhalerao, R., Groover, A. (2010). Genetics and genomics of Populus (New York: Springer).
Jiang, D., Feng, J., Dong, M., Wu, G., Mao, K., Liu, J. (2016). Genetic origin and composition of a natural hybrid poplar Populus x jrtyschensis from two distantly related species. BMC Plant Biol. 16, 89. doi: 10.1186/s12870-016-0776-6
Karhu, N., Hamet-Ahti, L. (1992). Gen. Populus. Suomen puu-ja pensaskasvio, (Helsinki: Dendrologian Seura), 142–152.
Kersten, B., Pakull, B., Groppe, K., Lueneburg, J., Fladung, M. (2014). The sex-linked region in Populus tremuloides Turesson 141 corresponds to a pericentromeric region of about two million base pairs on P. trichocarpa chromosome 19. Plant Biol. (Stuttg) 16 (2), 411–418. doi: 10.1111/plb.12048
Kim, G., Leite Montalvao, A. P., Kersten, B., Fladung, M., Müller, N. (2021). The genetic basis of sex determination in Populus provides molecular markers across the genus and indicates convergent evolution. Silvae Genetica 70, 145–155. doi: 10.2478/sg-2021-0012
Kobolkuti, Z. A., Cseke, K., Benke, A., Bader, M., Borovics, A., Nemeth, R. (2019). Allelic variation in candidate genes associated with wood properties of cultivated poplars (Populus). Biol. Futur. 70 (4), 286–294. doi: 10.1556/019.70.2019.32
Koltzenburg, M. (1999). Bestimmungsschlüssel für in Mitteleuropa heimische und kultivierte Pappelarten und -sorten (Populus spec.). (Göttingen: E. Goltze)
Kulagin, Y. Z. (1974). Woody plants and industrial environment [Drevesnye rasteniya i promyshlennaya sreda] (Moscow: Nauka).
Leskinen, E., Alström-Rapaport, C. (1999). Molecular phylogeny of Salicaceae and closely related Flacourtiaceae: evidence from 5.8 S, ITS 1 and ITS 2 of the rDNA. Plant Systematics Evol. 215, 209–227. doi: 10.1007/BF00984656
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997v2. doi: 10.48550/arXiv.1303.3997
Liu, X., Wang, Z., Shao, W., Ye, Z., Zhang, J. (2016). Phylogenetic and taxonomic status analyses of the abaso section from multiple nuclear genes and plastid fragments reveal new insights into the North America origin of Populus (Salicaceae). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.02022
Mayorov, S. R., Alekseev, Y. E., Bochkin, V. D., Nasimovich, Y. A., Shcherbakov, A. V. (2020). Alien flora of the Moscow Region: the composition, origin and the vectors of formation [Chuzherodnaya flora Moskovskogo regiona: sostav, proiskhozhdenie i puti formirovaniya] (Moscow: KMK).
Mayorov, S. R., Bochkin, V. D., Nasimovich, Y. A., Shcherbakov, A. V. (2012). Adventive flora of Moscow and Moscow Region [Adventivnaya flora Moskvy i Moskovskoi oblasti] (Moscow: KMK).
McCormack, J. E., Hird, S. M., Zellmer, A. J., Carstens, B. C., Brumfield, R. T. (2013). Applications of next-generation sequencing to phylogeography and phylogenetics. Mol. Phylogenet Evol. 66 (2), 526–538. doi: 10.1016/j.ympev.2011.12.007
McKown, A. D., Klapste, J., Guy, R. D., Soolanayakanahally, R. Y., La Mantia, J., Porth, I., et al. (2017). Sexual homomorphism in dioecious trees: extensive tests fail to detect sexual dimorphism in Populus (dagger). Sci. Rep. 7 (1), 1831. doi: 10.1038/s41598-017-01893-z
Melnikova, N. V., Dmitriev, A. A. (2022). Poplar - the source of clean air [Topolya - istochnik chisTogo vozduha]. Environ. Business 11, 120–123.
Melnikova, N. V., Kudryavtseva, A. V., Borkhert, E. V., Pushkova, E. N., Fedorova, M. S., Snezhkina, A. V., et al. (2019). Sex-specific polymorphism of MET1 and ARR17 genes in Populus x sibirica. Biochimie 162, 26–32. doi: 10.1016/j.biochi.2019.03.018
Melnikova, N. V., Pushkova, E. N., Dvorianinova, E. M., Beniaminov, A. D., Novakovskiy, R. O., Povkhova, L. V., et al. (2021). Genome Assembly and Sex-Determining Region of Male and Female Populus x sibirica. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.625416
Muller, N. A., Kersten, B., Leite Montalvao, A. P., Mahler, N., Bernhardsson, C., Brautigam, K., et al. (2020). A single gene underlies the dynamic evolution of poplar sex determination. Nat. Plants 6 (6), 630–637. doi: 10.1038/s41477-020-0672-9
Nasimovich, Y. A., Kostina, M. V., Vasilyeva, N. V. (2019). The concept of species in poplars (genus Populus L., Salicaceae) based on the example of the subgenus Tacamahaca (Spach) Penjkovsky representatives growing in Russia and neighbouring countries [Koncepciya vida u topolej (genus Populus L., Salicaceae) na primere predstavitelej podroda Tacamahaca (Spach) Penjkovsky]. Environ. Human: Ecol. Stud. 9 (4), 426–466. doi: 10.31862/2500-2961-2019-9-4-426-466
Nasimovich, Y. A., Vasilyeva, N. V. (2019). Comparison of morphological characters of different poplar species (Populus, Salicaceae) using the example of Russian and Central Asian Tacamahaca (Spach) Penjkovsky subgenus representatives [Sravnenie po morfologicheskim priznakam raznyh vidov topolej (Populus, Salicaceae) na primere rossijskih i sredneaziatskih predstavitelej podroda Tacamahaca (Spach) Penjkovsky]. Environ. Human: Ecol. Stud. 9 (3), 285–301. doi: 10.31862/2500-2961-2019-9-3-285-301
Navarro-Lopez, B., Granizo-Rodriguez, E., Palencia-Madrid, L., Raffone, C., Baeta, M., de Pancorbo, M. M. (2021). Phylogeographic review of Y chromosome haplogroups in Europe. Int. J. Legal Med. 135 (5), 1675–1684. doi: 10.1007/s00414-021-02644-6
Negi, M. S., Rajagopal, J., Chauhan, N., Cronn, R., Lakshmikumaran, M. (2002). Length and sequence heterogeneity in 5S rDNA of Populus deltoides. Genome 45 (6), 1181–1188. doi: 10.1139/g02-094
Novakovskiy, R. O., Dvorianinova, E. M., Rozhmina, T. A., Kudryavtseva, L. P., Gryzunov, A. A., Pushkova, E. N., et al. (2020). Data on genetic polymorphism of flax (Linum usitatissimum L.) pathogenic fungi of Fusarium, Colletotrichum, Aureobasidium, Septoria, and Melampsora genera. Data Brief 31, 105710. doi: 10.1016/j.dib.2020.105710
Page, R. D. (2000). Extracting species trees from complex gene trees: reconciled trees and vertebrate phylogeny. Mol. Phylogenet Evol. 14 (1), 89–106. doi: 10.1006/mpev.1999.0676
Pakull, B., Groppe, K., Mecucci, F., Gaudet, M., Sabatti, M., Fladung, M. (2011). Genetic mapping of linkage group XIX and identification of sex-linked SSR markers in a Populus tremula × Populus tremuloides cross. Can. J. For. Res. 41 (2), 245–253. doi: 10.1139/X10-206
Pakull, B., Groppe, K., Meyer, M., Markussen, T., Fladung, M. (2009). Genetic linkage mapping in aspen (Populus tremula L. and Populus tremuloides Michx.). Tree Genet. Genomes 5 (3), 505–515. doi: 10.1007/s11295-009-0204-2
Pakull, B., Kersten, B., Luneburg, J., Fladung, M. (2015). A simple PCR-based marker to determine sex in aspen. Plant Biol. (Stuttg) 17 (1), 256–261. doi: 10.1111/plb.12217
Paolucci, I., Gaudet, M., Jorge, V., Beritognolo, I., Terzoli, S., Kuzminsky, E., et al. (2010). Genetic linkage maps of Populus alba L. and comparative mapping analysis of sex determination across Populus species. Tree Genet. Genomes 6 (6), 863–875. doi: 10.1007/s11295-010-0297-7
Parker, K., Erzurumluoglu, A. M., Rodriguez, S. (2020). The Y chromosome: A complex locus for genetic analyses of complex human traits. Genes (Basel) 11 (11), 1273. doi: 10.3390/genes11111273
Povkhova, L. V., Pushkova, E. N., Rozhmina, T. A., Zhuchenko, A. A., Frykin, R. I., Novakovskiy, R. O., et al. (2022). Development and complex application of methods for the identification of mutations in the FAD3A and FAD3B genes resulting in the reduced content of linolenic acid in flax oil. Plants (Basel) 12 (1), 95. doi: 10.3390/plants12010095
Pushkova, E. N., Krasnov, G. S., Lakunina, V. A., Novakovskiy, R. O., Povkhova, L. V., Dvorianinova, E. M., et al. (2021). Genome and Transcriptome Sequencing of Populus x sibirica Identified Sex-Associated Allele-Specific Expression of the CLC Gene. Front. Genet. 12. doi: 10.3389/fgene.2021.676935
Regel, E. (1889). Russian dendrology [Russkaya dendrologiya] (St. Petersburg: Izdanie Karla Rikkera).
Roe, A. D., MacQuarrie, C. J., Gros-Louis, M. C., Simpson, J. D., Lamarche, J., Beardmore, T., et al. (2014). Fitness dynamics within a poplar hybrid zone: II. Impact of exotic sex on native poplars in an urban jungle. Ecol. Evol. 4 (10), 1876–1889. doi: 10.1002/ece3.1028
Sabatti, M., Gaudet, M., Muller, N. A., Kersten, B., Gaudiano, C., Scarascia Mugnozza, G., et al. (2020). Long-term study of a subdioecious Populus x canescens family reveals sex lability of females and reproduction behaviour of cosexual plants. Plant Reprod. 33 (1), 1–17. doi: 10.1007/s00497-019-00378-5
Schmidt, T., Schwarzacher, T., Heslop-Harrison, J. S. (1994). Physical mapping of rRNA genes by fluorescent in-situ hybridization and structural analysis of 5S rRNA genes and intergenic spacer sequences in sugar beet (Beta vulgaris). Theor. Appl. Genet. 88 (6-7), 629–636. doi: 10.1007/BF01253964
Schroeder, H., Hoeltken, A. M., Fladung, M. (2012). Differentiation of Populus species using chloroplast single nucleotide polymorphism (SNP) markers–essential for comprehensible and reliable poplar breeding. Plant Biol. (Stuttg) 14 (2), 374–381. doi: 10.1111/j.1438-8677.2011.00502.x
Skvortsov, A. K. (2008). About some poplars described by F.B. Fischer in 1841 [O nekotoryh topolyah, opisannyh F.B. Fisherom v 1841 g]. Bull. Main Botanical Garden 194, 61–67.
Skvortsov, A. K. (2010). Systematic synopsis of the genus Populus in Eastern Europe, Northern and Central Asia [Sistematicheskij konspekt roda Populus v vostochnoj Evrope, Severnoj i Srednej Azii]. Bull. Main Botanical Garden 196, 62–73.
Skvortsov, A. K., Belyanina, N. B. (2006). About balsam poplars (Populus, section Tacamahaca, Salicaceae) in the east of Asian Russia [O bal'zamicheskih topolyah (Populus, section Tacamahaca, Salicaceae) na vostoke aziatskoj Rossii]. Botanicheskii Zhurnal 91 (8), 1244–1252.
Slavov, G. T., Zhelev, P. (2010). “Salient biological features, systematics, and genetic variation of Populus,” in Genetics and genomics of Populus. Eds. Jansson, S., Bhalerao, R., Groover, A. (Berlin: Springer), 15–38.
Sokolov, S. Y., Shipchinsky, N. V., Yarmolenko, A. V. (1951). Genus Populus. Trees and shrubs of the USSR [Rod Topol'. Derev'ya i kustarniki SSSR] (Moscow - Leningrad: Akad. Nauk SSSR), 174–217.
Stettler, R. F., Zsuffa, L., Wu, R. (1996). “The role of hybridization in the genetic manipulation of Populus,” in Biology of Populus and its implications for management and conservation. Eds. Stettler, R. F., Bradshaw, H. D., Heilman, P. E., Hinckler, T. M. (Ottawa: Canadian Government Publishing), 87–112.
Suzuki, R., Shimodaira, H. (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22 (12), 1540–1542. doi: 10.1093/bioinformatics/btl117
Tsinovskis, R. (1977). “Two rare half-forgotten species of the genus Poplar (Populus L.) from the north-west of North America and related species and hybrids in Latvia,” in Botanicheskie sady pribaltiki. Okhrana rastenii. (Riga: Zinatne), 175–196.
Tsvelev, N. N. (2001). About poplars (Populus, Salicaceae) of St. Petersburg and Leningrad region [O topolyah (Populus, Salicaceae) Sankt-Peterburga i Leningradskoj oblasti]. Botanicheskii Zhurnal 86 (2), 70–78.
Wang, Z., Du, S., Dayanandan, S., Wang, D., Zeng, Y., Zhang, J. (2014). Phylogeny reconstruction and hybrid analysis of Populus (Salicaceae) based on nucleotide sequences of multiple single-copy nuclear genes and plastid fragments. PloS One 9 (8), e103645. doi: 10.1371/journal.pone.0103645
Wang, Y., Huang, J., Li, E., Xu, S., Zhan, Z., Zhang, X., et al. (2022). Phylogenomics and biogeography of Populus based on comprehensive sampling reveal deep-level relationships and multiple intercontinental dispersals. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.813177
Wang, D., Wang, Z., Kang, X., Zhang, J. (2019). Genetic analysis of admixture and hybrid patterns of Populus hopeiensis and P. tomentosa. Sci. Rep. 9 (1), 4821. doi: 10.1038/s41598-019-41320-z
Wang, M., Zhang, L., Zhang, Z., Li, M., Wang, D., Zhang, X., et al. (2020). Phylogenomics of the genus Populus reveals extensive interspecific gene flow and balancing selection. New Phytol. 225 (3), 1370–1382. doi: 10.1111/nph.16215
White, T. J., Bruns, T., Lee, S., Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: A guide to methods and applications. Eds. Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. (New York: Academic Press), 315–322.
Wilson, N. (2013). Genome analysis of Populus species: assessment of genetic diversity of P. deltoides, characterization of wide hybrids and phylogenetic analysis using molecular markers. Ph.D. Thesis (New Delhi: Teri University).
Wolkenstein, M. [P.W.] (1882). “New Plants at the Moscow Exhibition, in The gardeners' chronicle: a Weekly Illustrated. Journal of Horticulture and Allied Subjects. XVIII (New series. July to December), (London: Bradbury, Agnew, and Co.) 108.
Xin, H., Zhang, T., Wu, Y., Zhang, W., Zhang, P., Xi, M., et al. (2020). An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting. Plant J. 101 (2), 253–264. doi: 10.1111/tpj.14536
Xue, L., Wu, H., Chen, Y., Li, X., Hou, J., Lu, J., et al. (2020). Evidences for a role of two Y-specific genes in sex determination in Populus deltoides. Nat. Commun. 11 (1), 5893. doi: 10.1038/s41467-020-19559-2
Yang, W., Wang, D., Li, Y., Zhang, Z., Tong, S., Li, M., et al. (2021). A general model to explain repeated turnovers of sex determination in the salicaceae. Mol. Biol. Evol. 38 (3), 968–980. doi: 10.1093/molbev/msaa261
Yin, T., Difazio, S. P., Gunter, L. E., Zhang, X., Sewell, M. M., Woolbright, S. A., et al. (2008). Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res. 18 (3), 422–430. doi: 10.1101/gr.7076308
Zhang, S., Wu, Z., Ma, D., Zhai, J., Han, X., Jiang, Z., et al. (2022). Chromosome-scale assemblies of the male and female Populus euphratica genomes reveal the molecular basis of sex determination and sexual dimorphism. Commun. Biol. 5 (1), 1186. doi: 10.1038/s42003-022-04145-7
Zhou, R., Macaya-Sanz, D., Schmutz, J., Jenkins, J. W., Tuskan, G. A., DiFazio, S. P. (2020). Sequencing and analysis of the sex determination region of Populus trichocarpa. Genes (Basel) 11 (8), 843. doi: 10.3390/genes11080843
Keywords: Populus, Aigeiros, Tacamahaca, poplars, intersectional hybrids, molecular phylogeny, sex-determining region, targeted deep sequencing
Citation: Borkhert EV, Pushkova EN, Nasimovich YA, Kostina MV, Vasilieva NV, Murataev RA, Novakovskiy RO, Dvorianinova EM, Povkhova LV, Zhernova DA, Turba AA, Sigova EA, Snezhkina AV, Kudryavtseva AV, Bolsheva NL, Krasnov GS, Dmitriev AA and Melnikova NV (2023) Sex-determining region complements traditionally used in phylogenetic studies nuclear and chloroplast sequences in investigation of Aigeiros Duby and Tacamahaca Spach poplars (genus Populus L., Salicaceae). Front. Plant Sci. 14:1204899. doi: 10.3389/fpls.2023.1204899
Received: 12 April 2023; Accepted: 07 August 2023;
Published: 04 October 2023.
Edited by:
Satoshi Watanabe, Saga University, JapanReviewed by:
Konstantin V. Krutovsky, University of Göttingen, GermanySoichiro Nagano, Forestry and Forest Products Research Institute, Japan
Copyright © 2023 Borkhert, Pushkova, Nasimovich, Kostina, Vasilieva, Murataev, Novakovskiy, Dvorianinova, Povkhova, Zhernova, Turba, Sigova, Snezhkina, Kudryavtseva, Bolsheva, Krasnov, Dmitriev and Melnikova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nataliya V. Melnikova, mnv-4529264@yandex.ru
†These authors have contributed equally to this work
 Elena V. Borkhert
Elena V. Borkhert Elena N. Pushkova
Elena N. Pushkova Yuri A. Nasimovich2†
Yuri A. Nasimovich2† Natalia V. Vasilieva
Natalia V. Vasilieva Ramil A. Murataev
Ramil A. Murataev Roman O. Novakovskiy
Roman O. Novakovskiy Ekaterina M. Dvorianinova
Ekaterina M. Dvorianinova Liubov V. Povkhova
Liubov V. Povkhova Daiana A. Zhernova
Daiana A. Zhernova Anastasia A. Turba
Anastasia A. Turba Elizaveta A. Sigova
Elizaveta A. Sigova Anastasiya V. Snezhkina
Anastasiya V. Snezhkina Anna V. Kudryavtseva
Anna V. Kudryavtseva Nadezhda L. Bolsheva
Nadezhda L. Bolsheva George S. Krasnov
George S. Krasnov Alexey A. Dmitriev
Alexey A. Dmitriev Nataliya V. Melnikova
Nataliya V. Melnikova