- 1Key Laboratory of Grassland Resources of Ministry of Education, College of Grassland, Resources and Environment, Inner Mongolia Agricultural University, Hohhot, Inner Mongolia, China
- 2College of Geographical Science, Inner Mongolia Normal University, Hohhot, China
Land salinization considerably limits crop production. Biological improvement of saline and alkaline land is an important way to achieve efficient land use. It is crucial to study the salt tolerance of halophyte resources in order to explore and improve plant resources through biological improvement. Glaux maritima is a mesophyte halophyte with strong salt tolerance. In this study, we conducted research on the salt tolerance mechanism of G. maritima through phenotypic, physiological, and transcriptomic aspects. The results indicate that leaf cross-sections revealed that G. maritima has a salt gland tissue composed of stalk, collecting, and secretory cells, which are trapped in epidermal cells. At the physiological level, the maximum salt tolerance threshold of G. maritima leaves was 600 mM/L. At this concentration, proline content, relative conductivity, and superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) enzyme activities were maximum. At the transcriptional level, transcriptome data of three experimental groups (N0: 0 mM/L, N3: 600 mM/L, and N4: 800 mM/L) were analyzed, and six essential genes related to proline synthesis and five essential genes related to SOD and CAT enzyme activities were identified. Two genes involved in CAT enzyme activity were also found to play an important role in the MAPK signaling pathway. Trend analysis revealed that the MAPK signaling regulation (37 differentially expressed genes (DEGs)), phytohormone regulation (48 DEGs), glutathione metabolism (8 DEGs), flavonoid and flavonoid biosynthesis (2DEGs), and flavonoid biosynthesis (24 DEGs) pathways played important roles in regulating the salt tolerance of G. maritima. These findings provide valuable information for further studies on the functional characteristics of G. maritima in response to abiotic stress and may contribute to salt resistance breeding of fodder crops for cultivation in saline alkali land.
Introduction
The salinization of soil is a major abiotic stressor that inhibits plant development. It is estimated that 831 million hectares, or around 6% of the world’s total land area, are ruined by salt and cannot be utilized for agricultural (Shrivastava and Kumar, 2015; Kuang et al., 2019; Safdar et al., 2019). Intensifying salinization of soils is a result of both human activity and climate change, and this has devastating effects on plant development and agricultural yields in salinized regions (Munns et al., 2019). Using and improving arid and salty soil is greatly facilitated by halophytes. Increased land-use efficiency, reduced soil erosion, and remediation of soil salinity are all possible due to halophytes, which can develop regularly despite harsh environmental circumstances including high salt and drought. Studies have shown that wild species of the 5 genera Artemisia vulgaris L., Artemisia absinthium L., Chenopodium album L., Sanguisorba minor Scop. and Salsola komarovii Iljin. can not only grow normally on land affected by different degrees of salinization, but can also be used to remediate soil salinity and heavy metals (Rebhi et al., 2019; Calone et al., 2021; Trofimova et al., 2012; Walker et al., 2014; Saddhe et al., 2020; Tlili et al., 2020).
Adaptation of halophytes to salt stress is reflected in their morphology and physiology. Halophytes often have a unique salt-tolerant structure that resists salt damage. Studies have shown that the unique structures of salt-secreting plants are mainly salt glands and salt vesicles (Wu et al., 2020). Salt glands are commonly found in monocotyledons, such as Aeluropus, Sporobolus, Spartina, and Zoysia of the Gramineae family, and flowering dicotyledons in the desert and coastal habitats, such as Avicenia, Tamarix, Reaumuria Linn., and Limonium Mill. These plants contain multicellular salt glands with different structures and cell numbers (Zhou et al., 2001; Wilson et al., 2017). Salt vesicles are mainly found in Atriplex, Chenopodium, and Salsola (Zhou et al., 2001). They are salt-secreting tissues that store salt first and discharge it after the salt vesicles mature and rupture. These tissues are composed of vesicle cells (EBCs), stalk cells (SCs), and epidermal cells (ECs) (Shabala et al., 2014). Physiologically, halophyte seedlings resist salt damage by the regulation of ionic, osmotic, and oxidative systems (Yuan et al., 2015). After salt stress, plant membrane lipid peroxidation decomposes into malondialdehyde, which destroys the structure and function of the plasma membranes, changes the permeability of the membrane, and affects the normal physiological and biochemical reactions of plants. Simultaneously, plant cytoplasm accumulates large amounts of proline, maintains intracellular homeostasis, and reduces ionic toxicity (Shabala et al., 2014). Antioxidants such as carotenoids and glutathione, which are produced by plants under oxidative stress, can trigger a large amount of antioxidants, especially ascorbic acid and glutathione. Plants respond to potentially damaging oxidative stress by turning on redox regulating enzymes like glutathione reductase (GR) and antioxidant enzyme systems including superoxide dismutase (SOD), and ascorbic acid peroxidase (APX), and catalase (CAT), among others (Yuan et al., 2012). In a study of the salt-tolerant growth strategy of the halophyte Salicornia europaea, it was found that, after salt stress, while biomass and chlorophyll content decrease, proline content and peroxidase activity increase (Ghanem et al., 2021). In saline-alkali lands, halophytes balance the osmotic pressure inside and outside the cells through their unique salt glands or salt vesicles. At the same time, the content of physiological and biochemical substances in the plant changes, affecting the osmotic pressure inside and outside plant cells and the permeability of the cell membrane, enabling halophytes to maintain normal growth in saline environments.
In order to discover the genes responsible for salt tolerance in halophytes, high-throughput transcriptome sequencing has been frequently employed. Zhao et al. (Zhao et al., 2014). mapped the transcriptome of Salicornia paniculata (Chenopodiaceae) and discovered that genes involved in glucanase, 6-phosphate dehydrogenase, aldehyde dehydrogenase, ethylene response factor, MYB, WRKY, and bZIP transcription factor, citrate synthase, and protein kinase play a significant role in plant salt tolerance under 600 mM NaCl stress. Transcriptome analysis of Zoysia japonica exposed to salt stress using RNA-seq technology has also been performed. The majority of DEGs show enrichment in GO functional categories including stimulation and stress responses (Xie et al., 2015). Transcriptome analysis of Suaeda glauca under salt stress showed that differential genes are significantly enriched in functional clusters, such as signal transduction, transporters, cell wall, and growth (Jin et al., 2016). Analysis of transcriptomes of S. paniculata under salt stress revealed the expression of the DNA damage repair-related genes hcrev1 and hcrev3 induced by salt stress (Du et al., 2017).
Inner Mongolia is located in northern China, which is a typical inland area and the climate is drought and less rain. As an important agricultural and animal husbandry crop producing area in China, the planting mode is mainly irrigation, leading to an increasing degree of secondary salinization in the soil year by year. There is an urgent need to explore salt tolerant mesophytes that are suitable for the ecological environment of the region and cultivate agricultural and pastoral crops with good salt tolerance. Glaux maritima L. is a plant of Glaux Linn. in Primulaceae, and is a mesophytic halophyte. Widely distributed in the western and northern regions of China. Although previous studies have conducted preliminary studies on the salt tolerance of G. maritima from aspects such as ion content and the ecology of community microorganisms under salt stress, there are still few reports on the salt tolerance adaptation strategies and molecular mechanisms of G. maritima. (Rozema, 1975; Rozema and Sminia, 1977; Yamamoto et al., 2020; Yamamoto et al., 2021). Therefore, this study conducted research on the morphological structure, physiological metabolism and Transcriptome of G. maritima. seedlings under varying salt concentrations. To provide theoretical reference for later salt tolerant gene mining and provide new resource support for improving salt tolerance of agricultural and livestock crops, and provides guidance for improving the salt tolerance of crops and their cultivation in saline-alkali areas.
Materials and methods
Inner Mongolia Agricultural University’s Key Laboratory of Grassland and Resources of the Ministry of Education cultivated wild G. maritima seedlings in 1/2 Hoagland solution in a greenhouse for 15 days (wild G. maritima seedlings were collected from the Hailiutu base of Inner Mongolia Agricultural University Science and Technology Park, Bikeqi Town, Tumet Left Banner, Hohhot, Inner Mongolia, N:40° 38 ′, E:111° 28 ′, Height:1060 m.). The optimal growing circumstances included a steady 25°C temperature, 60% relative humidity, and 1600 lx of light. Selected G. maritima seedlings were subjected to NaCl concentrations of 0, 200, 400, 600, and 800 mM during 24 hours. Malondialdehyde (MDA), Pro, conductivity, and the activity of superoxide dismutase (SOD), peroxide (POD), glutathione (GSH) reductase (GPX), and catalase (CAT) were measured in G. maritima leaf tissues of 0.2 g Each index measurement was repeated thrice for each of the five treatments (0, 200, 400, 600, and 800 mM NaCl).
Paraffin sections
Fresh leaf tissue of G. maritima was fixed with FAA fixative (formaldehyde: glacial acetic acid: ethanol = 0.5:0.5:9) for > 24 h. The blades and labels were placed in the fume hood into the dehydration tank for dehydration and wax soaking. After being dried out, samples were subjected to various chemical treatments, including 4 hours in 75% ethanol, 2 hours in 85% ethanol, 2 hours in 90% ethanol, 1 hour in 95% ethanol, 30 minutes in anhydrous ethanol I, and ethanol II, 5-10 minutes in alcohol benzene, and 5-10 minutes in xylene II. The wax was soaked for an hour in paraffin I, II, and III, all melted at 65° C. After embedding, using a paraffin microtome, thin slices (4 μm) were cut from the samples. Finally, the slices were flattened in 40 °C warm water, dried in a 60 °C oven, and then retrieved and stored at room temperature. Later, an optical microscope was used to examine the cells (Liu, 2006).
Determination of MDA, proline, and relative conductivity
Fresh plant leaf tissue (0.2 g) was homogenized at 4°C in 5% (w/v) trichloroacetic acid (TCA) (10 mL) to determine malondialdehyde (MDA). Two ml of the supernatant and two ml of 0.67 percent (m/v) thiobarbituric acid (prepared in 10% TCA) were added to the homogenate after centrifugation at 4000 rpm for 10 minutes. The samples were heated to 95°C for 20 minutes before being cooled to 25°C in an ice bath. After re-centrifuging the samples for 5 minutes at 4000 rpm, the absorbance at 450, 532, and 600 nm was measured (Khan et al., 2021). To calculate the MDA concentration, the absorption coefficient was used i.e.,
To calculate proline concentration, we homogenized 0.2 g of fresh plant leaf tissue in 3.0% sulphosalicylic acid (2.5 ml). The homogenate was centrifuged for 10 minutes at 1000 rpm/min. Two ml of the supernatant, two ml of glacial acetic acid, and three ml of acid ninhydrin were placed in a test tube. The mixture was heated for 30 minutes in a 100°C water bath before being cooled rapidly in an ice bath. Initially, the solution was chilled, and afterwards, 4 mL of toluene was poured prior to vortexed. A fresh test tube was used for the toluene (top layer) containing the chromophore. In the end, absorbance at 520 nm was determined using toluene as a standard in a spectrophotometer. As a result of employing a standard curve, the proline content was calculated and reported in mg·g-1 (Khan et al., 2021).
20 new leaves were chosen at random, trimmed with scissors, added to 20 ml of deionized water, and let to soak for 24 hours before the conductivity of the resulting solution was recorded as R1. They were given a 10-minute boil-water bath before being allowed to remain at room temperature before another conductivity measurement was taken and the result was recorded as R2. The following equation was used to get the relative conductivity: (Ma et al., 2003)
Determination of SOD, POD, CAT, and APX enzyme activity
Liquid nitrogen was used to crush fresh leaves (0.2 g), which were then dissolved in PBS and centrifuged for 10 minutes at 4°C at 12000 rpm/min. The antioxidant enzymes superoxide dismutase (SOD), catalase (CAT), and ascorbic acid peroxidase (APX) were tested using a nitroblue tetrazolium (NBT) light, dioxygen water, and ascorbic acid reduction method (APX) (McCord and Fridovich, 1969; Beers and Sizer, 1952; Chance and Maehly, 1955; Nakano and Asada, 1981).
RNA isolation
Total RNA was extracted using a TRIzol reagent kit (Invitrogen, Carlsbad, CA, USA), as per the manufacturer’s instructions (Pandey et al., 2022). To verify the integrity of the RNA, we processed it through an RNase-free agarose gel electrophoresis and then analyzed the results using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Following total RNA isolation, While Oligo(dT) beads were used to remove rRNA from eukaryotic mRNA, the Ribo-ZeroTM Magnetic Kit was used to do the same for bacterial mRNA (Epicenter, Madison, WI, USA) (Attached Figure 1, Supplementary Table 1). The enriched mRNA was fragmented into tiny bits using fragmentation buffer before being used for reverse transcription into cDNA. Second-strand cDNA was synthesized using DNA polymerase I, deoxynucleoside triphosphates, RNase H, and a buffer. Furthermore, the extraction of cDNA was carried out via QiaQuick PCR extraction kit (Qiagen, Venlo, The Netherlands), then it was end-repaired, had an extra base added, and finally, Illumina sequencing adapters were utilized to ligate it. Finally, Size-selected agarose gel electrophoresis was used to isolate the ligation products, which were then amplified by PCR and sequenced on an Illumina Novaseq 6000. (Gene Denovo Biotechnology, Guangzhou, China).
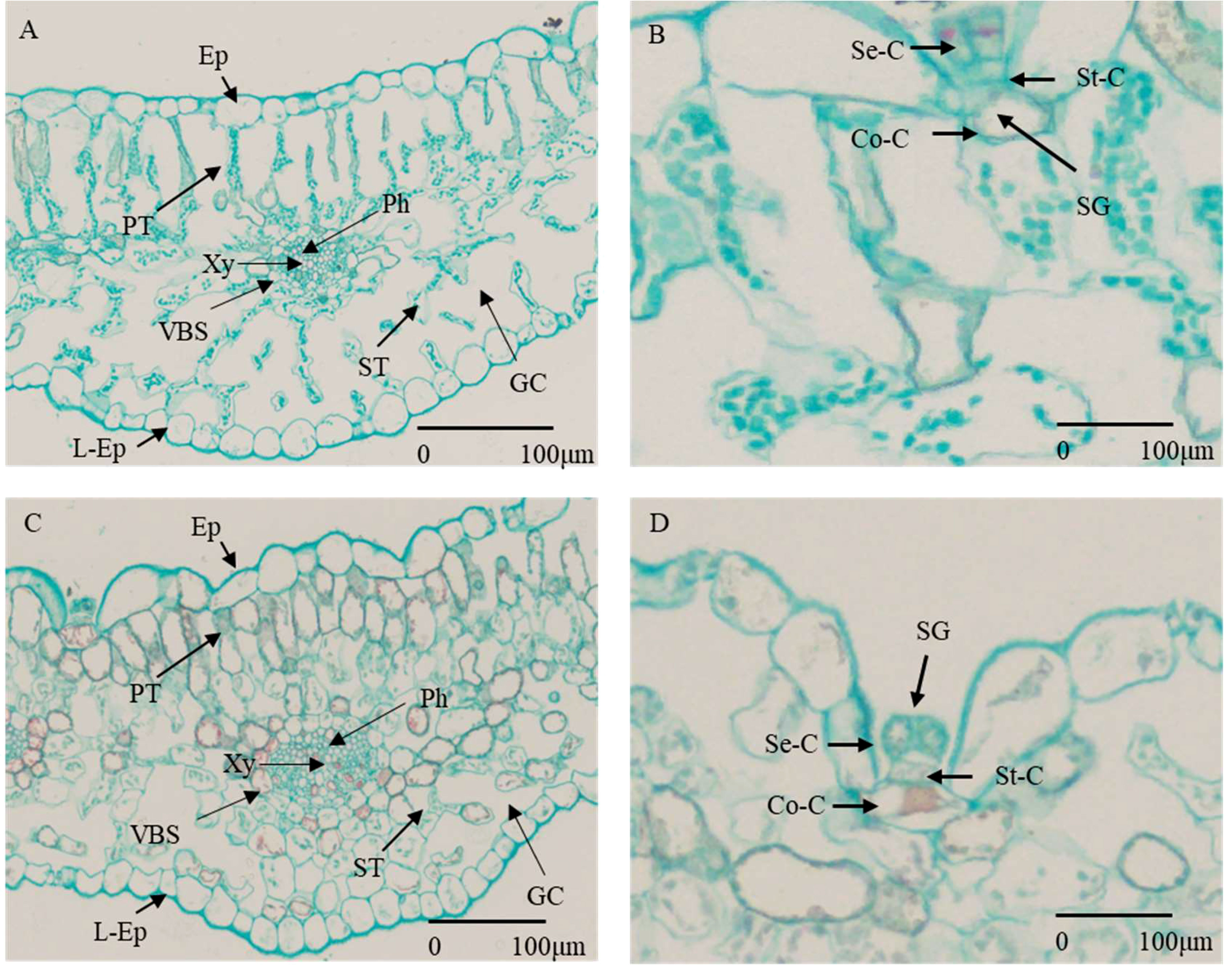
Figure 1 Cross section of G. maritima leaves under different salt concentrations. (A) State of various cells under 0 mM/L NaCl stress. (Ep, epidermis; L-Ep, lower epidermis; PT, palisade tissue; Ph, phloem; Xy, xylem; VBS, vascular bundle sheath; ST, spongy tissue; GC, gas chamber.) (B) Composition and status of salt glands under 0 mM/L NaCl stress (SG, salt gland; Se-C, secretory cell; St-C, stalk cell; Co-C, collecting cells.) (C) State of various cells under 800 mM/L NaCl stress (abbreviations are the same as for A). (D) Composition and status of salt glands under 800 mM/L NaCl stress (abbreviations are the same as for B).
Data analysis
Excel 2019 was used for preliminary data processing. Several variables were analyzed using univariate analysis with SPSS (version 19.0; SPSS, Chicago, Illinois, USA) to demonstrate their importance across a range of salt stress treatments, and the differential gene statistics were based on the results of differential analysis. The screening of significantly DEG was based on | log2FC|>1 and FDR < 0.05. The expression patterns of the gene were clustered using trend analysis, followed by analysis using the short time series expression miner tool, then enrichment using KEGG and GO terms. In order to get the enriched GO keywords and pathways, we used a threshold of Q ≤ 0.05.
qRT−PCR verification
TransScript One-Step gDNA Removal and Cdna Synthesis SuperMIX was used to reverse transcribe RNA. PerfectStart Green qPCR SuperMIX was used, which contained a 20 μL solution (50 ng cDNA as a template;10 × ROX reference dye), for qRT-PCR in a LightCycler 480 (Roche). This was conducted on each sample. The amplification protocol was as follows: 94°C for 30 sec, then 40 cycles at 94°C for 5sec, 60°C for 34sec and 72°C for 10 s, and then melt curve at 94°C 15 s, 60°C 1 min, and 94°C 15 s. Supplementary Table 4 lists the sequence of the specific primers used for qRT-PCR. β-Actin was used as an internal control. All qRT-PCR analyses were performed in three replicates. The relative gene expression levels were calculated using the 2−ΔΔCt method.
Results
Plant morphological changes under salt stress
The observation of changes in plant phenotype under abiotic stress was an important indicator for evaluating plant tolerance. By observing the transverse section of the plant leaf tissue, we can better observe the salt-tolerant structure of halophytes (Zhao, 2016). As a salt-secreting halophyte, with the increase of salt stress concentration, the phenotype of G. maritima seedlings were changed (Figures 2A–E). The leaves of G. maritima seedlings curled and salted out under 800 mM NaCl stress (Figure 2E). When observing the cell structure of the leaves of G. maritima (Figures 1A–D), it was found that the salt glands were the main salt-excreting tissue. The tissue was trapped in EC, which were composed of stalk cells (ST-Cs), collecting cells (Co-Cs), and secretory cells (SE-Cs) (Figure 1B, D).

Figure 2 Changes in G. maritima leaves after 24 h under salt stress. (A–E) Leaf morphology of G. maritima under 0, 200, 400, 600, and 800 mM/L NaCl stress. The small picture in the lower left corner shows the salt secretion state of a single leaf.
The thickness of leaf, the epidermis and lower epidermis, palisade tissue, and spongy tissue, as well as the size of the air cavity, all decreased in G. maritima leaves as the salt concentration increased (Figure 3A–D, F). The thickness of the upper and lower EC, as well as the size of the air cavity, were substantially decreased by 13.31%, 8.93%, and 30.81% and 14.94%, 15.68%, and 38.61%, after being treated to 200 and 400 mM NaCl, respectively, compared to control (Figure 3B, C, F, P < 0.05). Under treatment with 600 and 800 mM NaCl, the leaf thickness decreased by 33.90% and 39.76%, respectively, compared to the control. Upper EC thickness was decreased by 22.06% and 23.33% (P < 0.05) with reference to the control group (Figure 3B), the thickness of the lower EC decreased significantly by 23.99% and 24.64% (Figure 3C), the thickness of the palisade tissue decreased significantly by 38.63% and 40.74% (Figure 3D), and the thickness of the spongy tissue decreased by 36.36% and 51.50%, respectively (Figure 3E). Compared to the control treatment, the cavity area decreased the most (56.20% and 80.28%, respectively) (Figure 3F).
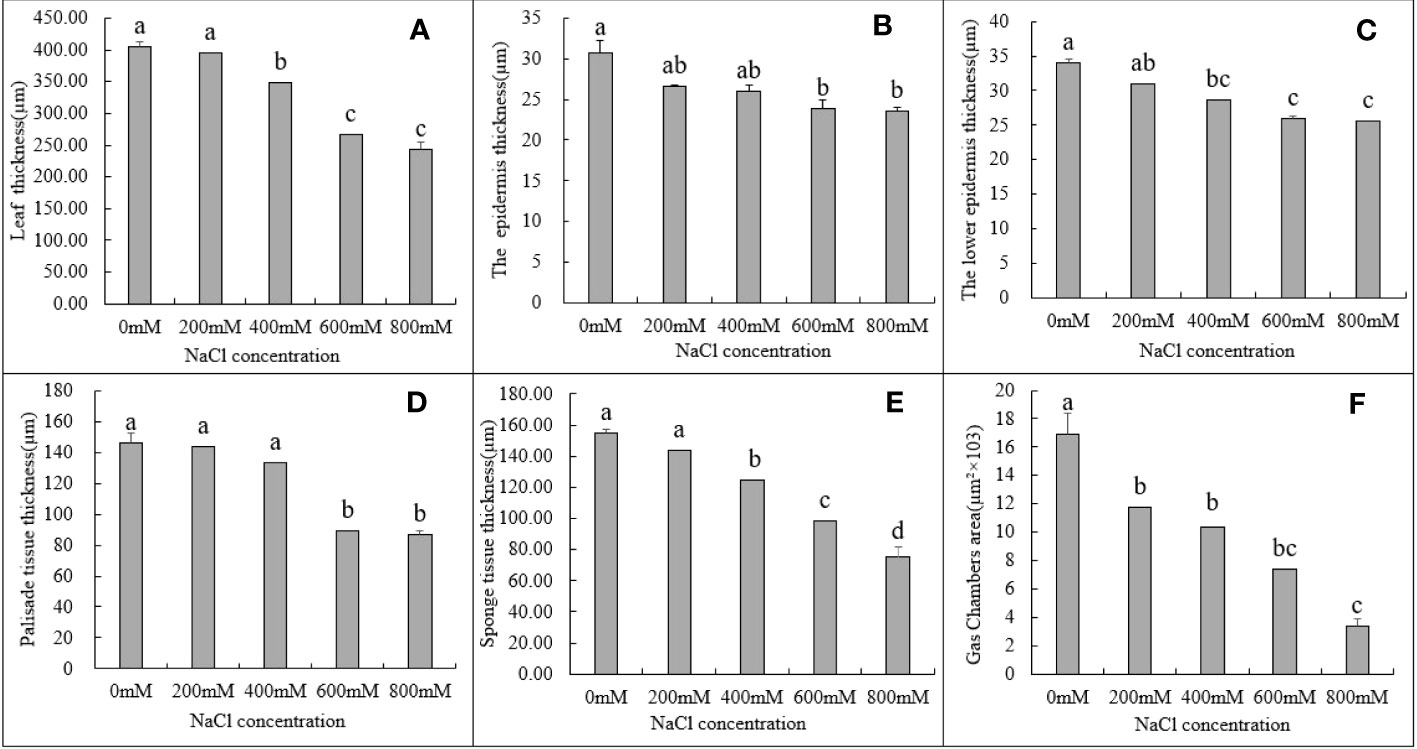
Figure 3 Changes in various tissues in the leaves of G. maritima under different salt concentrations stress. (A) The thickness in the leaves of G. maritima under different salt concentrations stress; (B, C) The epidermis and lower epidermis in the leaves of G. maritima under different salt concentrations stress; (D) Palisade tissue in the leaves of G. maritima under different salt concentrations stress; (E) Sponge tissue in the leaves of G. maritima under different salt concentrations stress; (F) Gas chamber area in the leaves of G. maritima under different salt concentrations stress. Different letters indicate significant differences under different salt concentrations (P < 0.05).
Physiological changes under salt stress
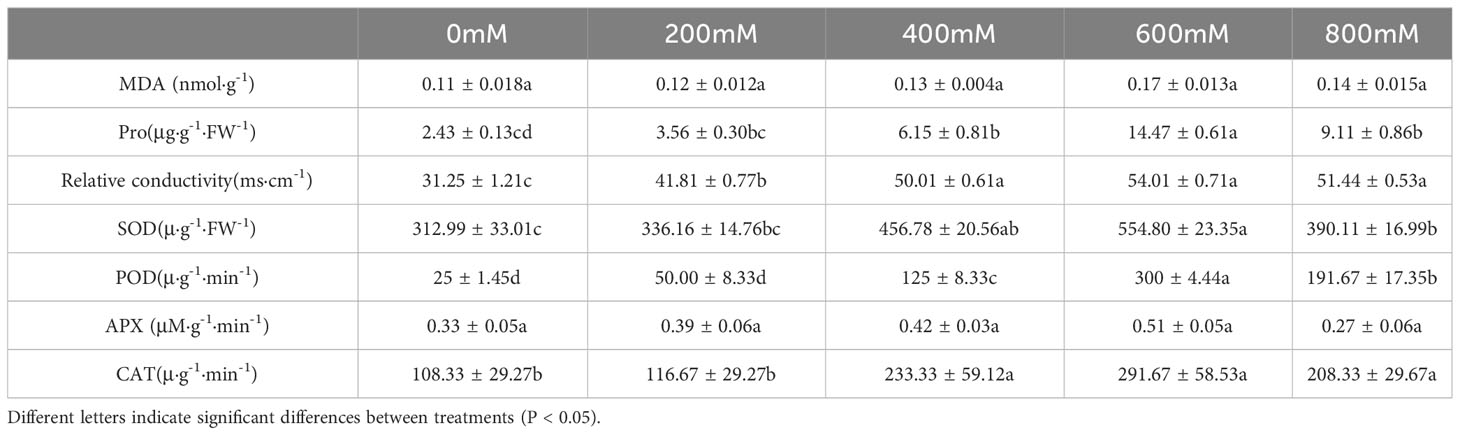
In addition to the phenotypic changes observed in plants under salt stress, ROS, malondialdehyde, and cell membranes in plants initiate corresponding tolerance mechanisms (Gong et al., 2014; Zou et al., 2020). Physiological indicators such as proline and malondialdehyde contents and plant antioxidant enzyme activity may be used to evaluate the salt tolerance mechanisms of G. maritima. Proline concentration was highest under 600 mM NaCl stress. Up to 600 mM NaCl, activity was significantly higher, however, they were lower at 800 mM NaCl, and proline content significantly increased in the leaves of G. maritima in all NaCl treatments in comparison to the control group (p < 0.05; Figure 4A, Table 1).
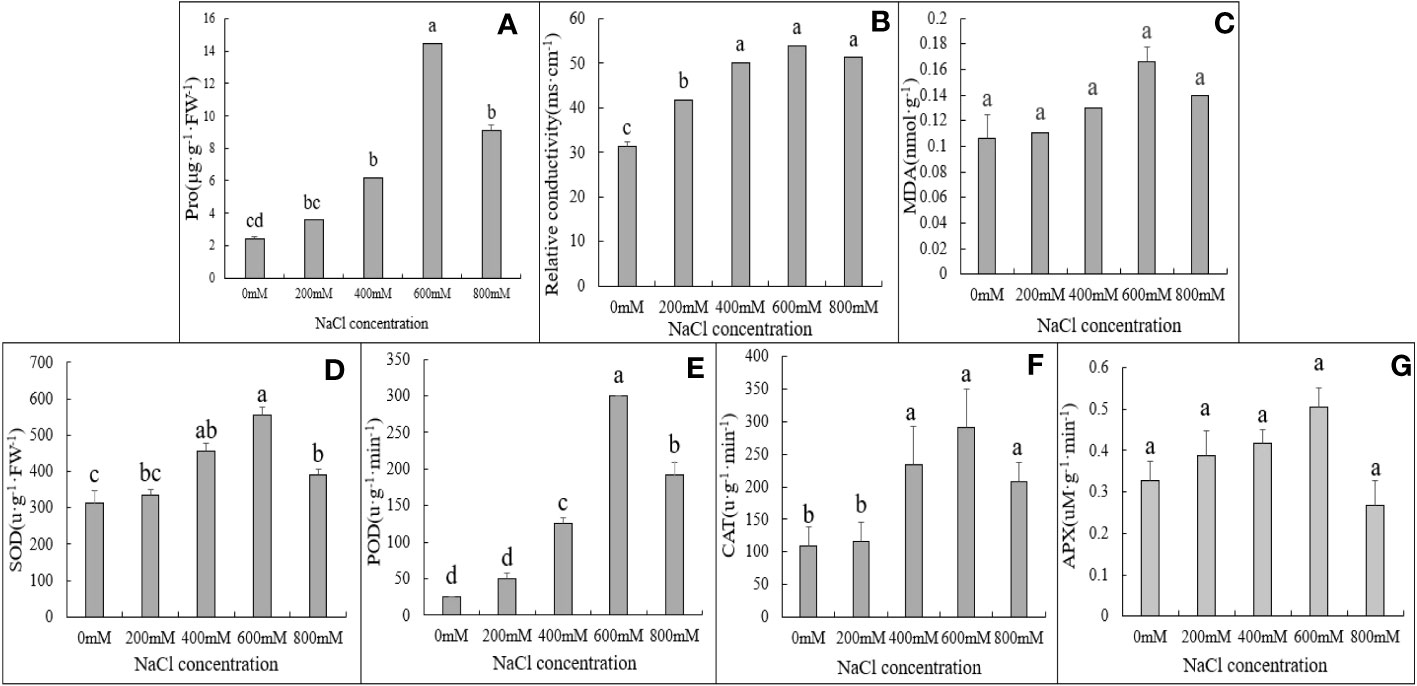
Figure 4 Physiological and biochemical indexes of G. maritima leaves under salt stress. (A) proline content of G. maritima leaves under salt stress; (B) Relative conductivity of G. maritima leaves under salt stress; (C) MDA of G. maritima leaves under salt stress; (D) SOD enzyme activity of G. maritima leaves under salt stress; (E) POD enzyme activity of G. maritima leaves under salt stress; (F) CAT enzyme activity of G. maritima leaves under salt stress; (G) APX enzyme activity of G. maritima leaves under salt stress. Different letters indicate significant differences under different salt concentrations (P < 0.05).
Relative conductivity analysis revealed that salt stress substantially increased lipid peroxidation in G. maritima leaves (p < 0.05). In the leaves of G. maritima, relative conductivity rose up to 600 mM NaCl in comparison to the control and remained noticeably high under all salt treatments (p < 0.05; Figure 4B, Table 1). The MDA concentration of G. maritima did not change between the salt stress treatments and the control, however (p> 0.05; Figure 4C, Table 1). Under various NaCl treatments, a similar trend was seen for the POD and SOD activity. Up to 600 mM NaCl, SOD and POD activity were significantly higher than control (p < 0.05), however they were lower at 800 mM NaCl. To a concentration of 600 mM NaCl, interestingly, POD activity was 12 times greater than the control and SOD activity was 1.8 times higher (Figure 4D, E, Table 1).
Under high salt concentration stress (400-800mM NaCl), the CAT enzyme activity of G. maritima seedlings was significantly higher than that of the control (p<0.05). And at 600 mM NaCl, CAT activity reached its maximum, which increased by 1.69 times compared to the control (Figure 4F, Table 1). As the salt concentration increased, there was no significant difference in APX activity among different treatments(p>0.05) (Figure 4G, Table 1).
Transcriptome sequencing in G. maritima under salt stress
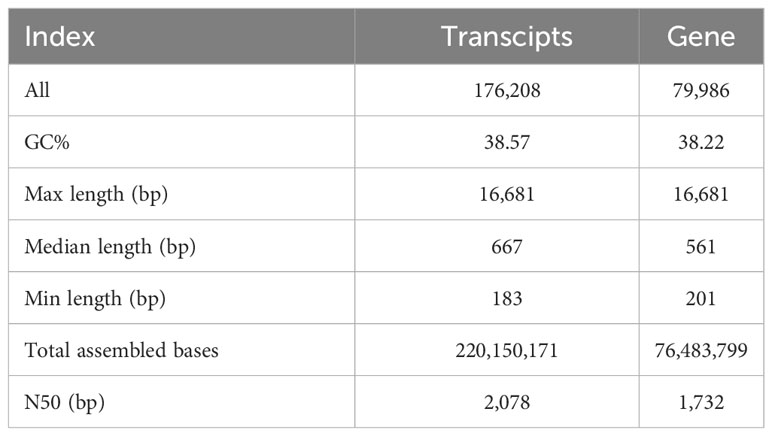
Based on the proline content and SOD and POD activities after different treatments, fresh G. maritima leaves after 24 h of 600 and 800mM NaCl treatments were collected; 0mM NaCl was used as the control. The RNA-seq experiment included nine libraries, one for each of the three replicates of each sample (N0: control; N3: plants under 600mM NaCl stress; and N4: plants under 800 mM NaCl stress). Subsequently, Total raw data sequences ranged from 36,348,544 to 49,426,720, yielding 6.23–8.48 GB of sequencing data (Supplementary Table 2). All the cDNA libraries yielded a total of 176,208 transcripts. After ensuring their integrity, the transcripts were combined into 79,986 unigenes with a N50 of 1,732 bp. Table 2 provides a brief overview of the transcriptome sequencing of G. maritima (Table 2).
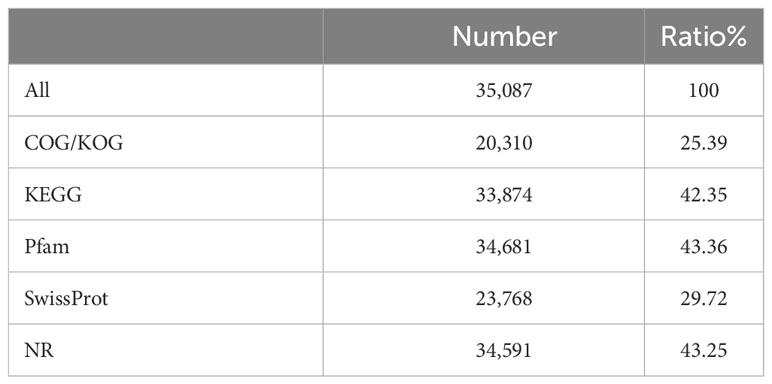
All assembled unigene clusters were aligned using BLASTX with an E-value cutoff of 0.00001 against orthologous group clusters for eukaryotic complete genomes SwissProt database (http://www.expasy.ch/sprot/), KEGG (http://www.genome.jp/kegg/), (GOG/KOG, (http://www.geneontology.org), Pfam database (http://pfam.xfam.org/), evolutionary genealogy of genes, and NCBI non-redundant protein database (NCBI_NR, http://www.ncbi.nlm.nih.gov/). The statistical findings from the six reputable databases are shown in Table 3.
Differentially expressed genes in G. maritima under salt stress
Differentially regulated genes in G. maritima in response to salt stress were found by comparing three sets of experimental conditions (N0, N3, and N4). Ten thousand eight hundred and nine genes’ expressions were found to be substantially varied in the three groups. Specifically, the comparisons between N0 and N3, N0 and N4, and N3 and N4 yielded 8285, 6637, and 402 DEGs, respectively (Figure 5A). Figure 5B presents a Venn diagram showing the overlap between the DEGs from the three comparisons. Between N0 and N4, there were 4,919 DEGs of overlap. Thirty-seven differentially expressed genes (DEGs) were found to be affected by therapy (Figure 5B). Table 4 displays data pertaining to the 37 DEGs throughout the three analyses (Table 4).
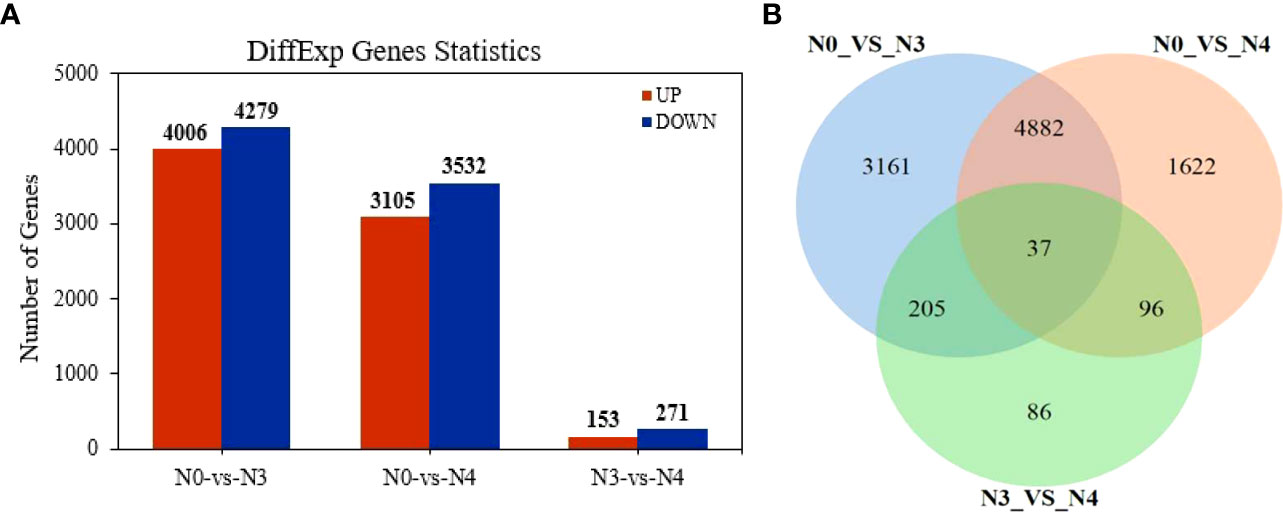
Figure 5 Differentially expressed genes in transcriptome data of G. maritima leaves under salt stress. (A) Statistics of differential genes after comparison at three concentrations; (B) Venn diagram of differential genes after comparison at three concentrations. N0:0 mM/L NaCl stress; N3:600mM/L NaCl stress; N4:800mM/L NaCl stress.
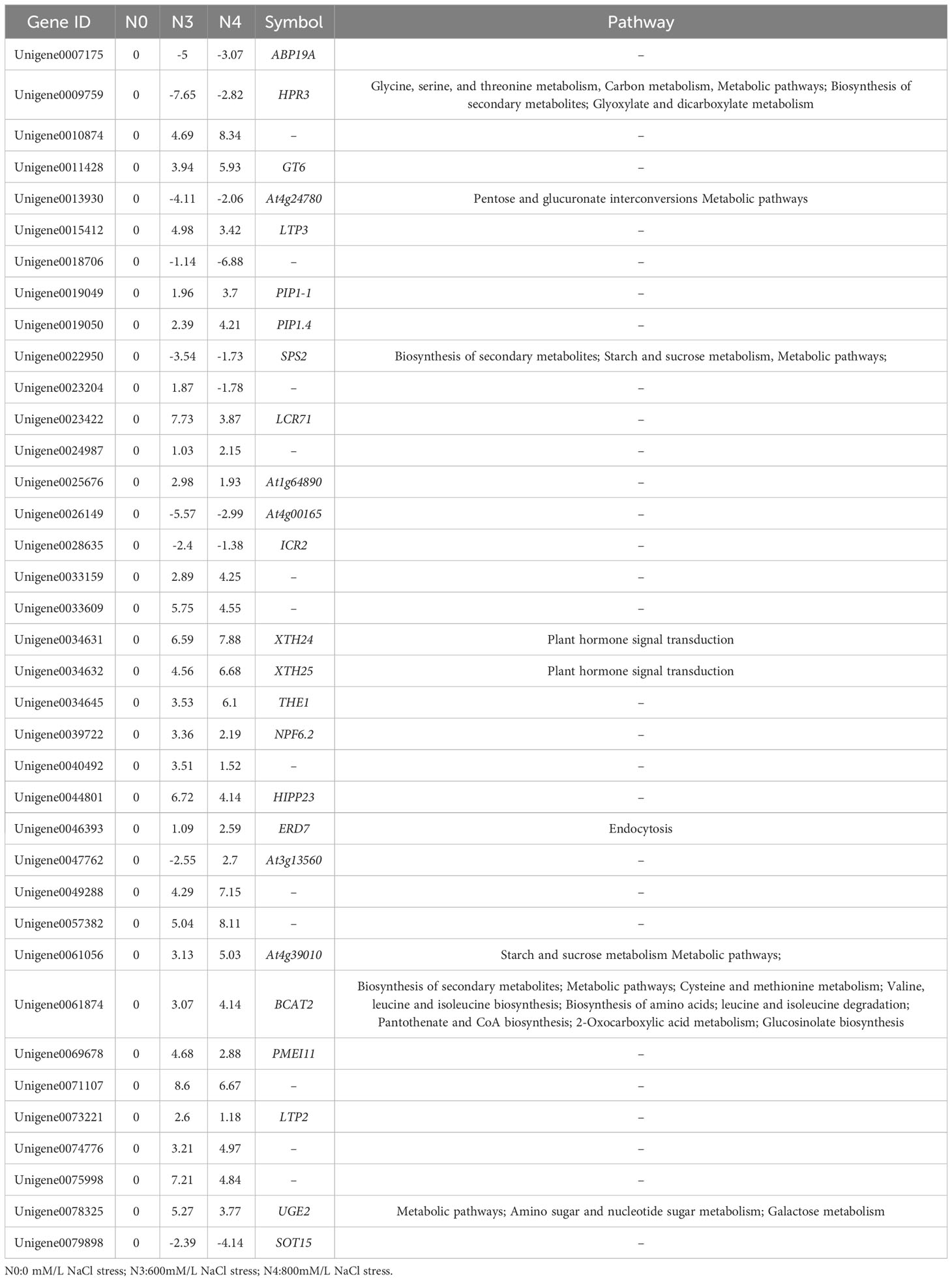
Table 4 Comparison of the N0 vs. N3, N0 vs. N4, and N3 vs. N4 groups regarding common differential gene ID, differential multiple, and participating pathways.
Gene expression under different salt stress treatments
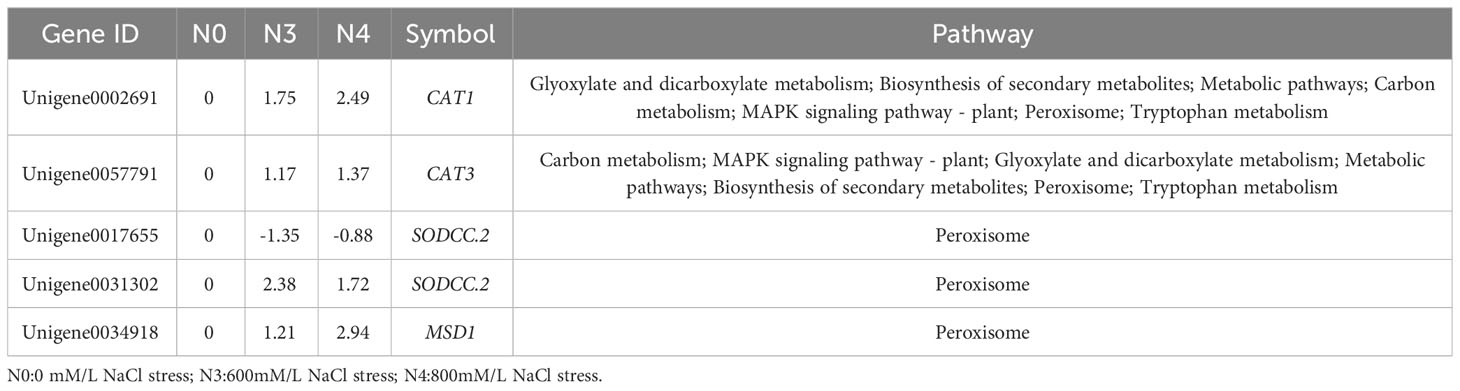
“Arginine and proline metabolism” and “peroxisome” were the primary key pathways identified. In the “arginine and proline metabolism” pathways, L-glutamate-5-semialdehyde encoding genes (δ-oat, Unigene0003065; PRO, Unigene0020312, Unigene005477, and Unigene0074521) all saw increases in their expression levels. One gene encoding L-glutamate-5-semialdehyde and L-glutamyl-phosphate also had its expression reduced in response to 800 mM NaCl stress (PRO2, Unigene0061751). Another downregulated gene encodes enhanced 1-pyrroline-5-carboxylate, which is involved in proline biosynthesis (POX2, Unigene0011171) (Figure 6, Table 5).
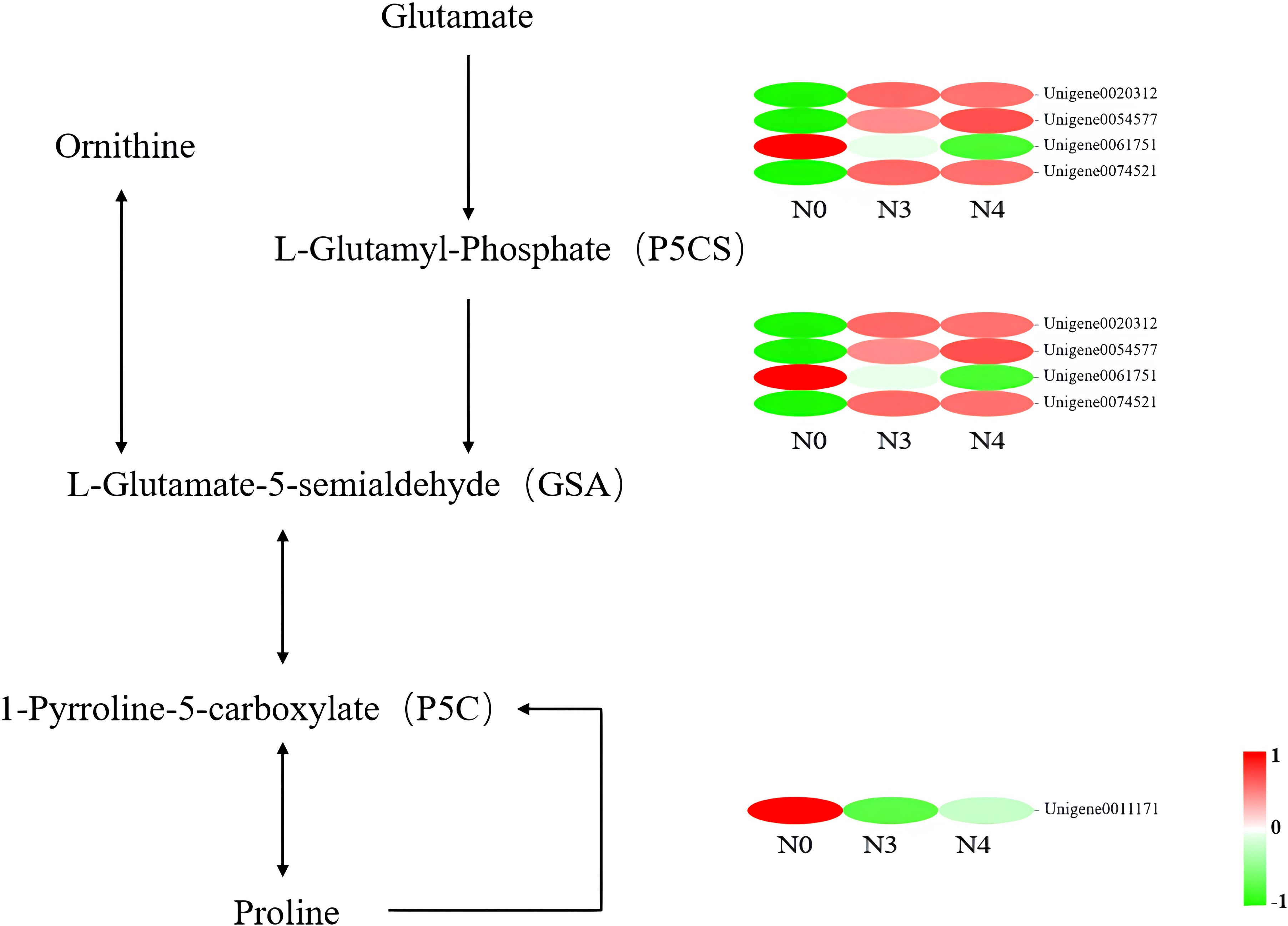
Figure 6 Heat map of differential gene expression in the arginine and proline metabolic pathways. N0:0 mM/L NaCl stress; N3:600mM/L NaCl stress; N4:800mM/L NaCl stress.
Genes related to peroxisomes under salt stress conditions were further investigated. In the “peroxisome” pathway, five genes affected SOD and CAT activities. These genes mainly affected the PTS1 type hydrogen peroxide metabolism in the antioxidant system of peroxisomal proteins. The expression levels of two genes encoding CAT were upregulated. These genes also affected H2O2 synthesis in the “MAPK signaling pathway – plant” pathway. Three negatively regulated genes encode SOD (MSD1, Unigene0034918) and SODCC.2 (Unigene0017655, Unigene0031302). Except for SODCC.2 (Unigene0017655), which was downregulated, the other two genes were upregulated (Table 6).
Trends in DEGs under different levels of salt stress
DEG expression patterns were identified using trend analysis by comparing DEGs at 24 h under varying NaCl concentrations. Most DEGs were enriched in profiles 0, 1, 6, and 7 when G. maritima was exposed to salt stress (P< 0.05). (Figure 7). KEGG analysis showed a distinct upward trend in expression (Figure 8), with profiles 7 and 6 showing considerable enrichment of DEGs. Critical pathways involved in disease resistance were also significantly enhanced, including “MAPK signaling pathway-plant” (37 DEGs), “glutathione metabolism” (8 DEGs), and “plant hormone signal transduction” (48 DEGs) (Figures 8C, D). Similar attention was paid to profile 0 since DEGs enriched there showed a distinct downregulated expression pattern (Figure 7). In addition, 4 profiles indicated enrichment for “flavone and flavanol biosynthesis” (2 DEGs) and “flavonoid biosynthesis” (24 DEGs), whose metabolites affect abiotic stress response (Figures 8A, B).
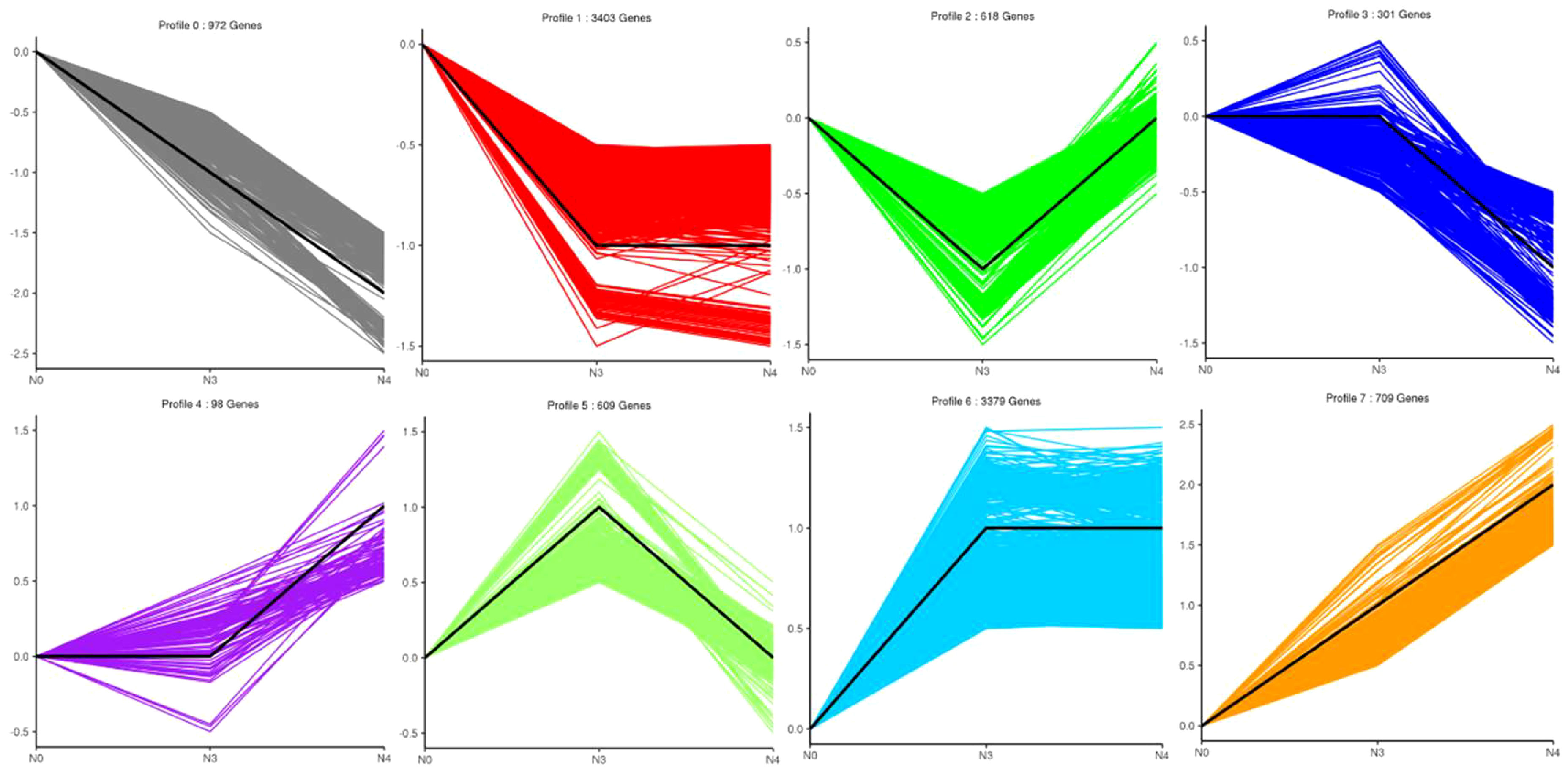
Figure 7 Number and trend of genes counted after trend analysis of transcriptome data of G. maritima after salt stress. N0:0 mM/L NaCl stress; N3:600mM/L NaCl stress; N4:800mM/L NaCl stress.
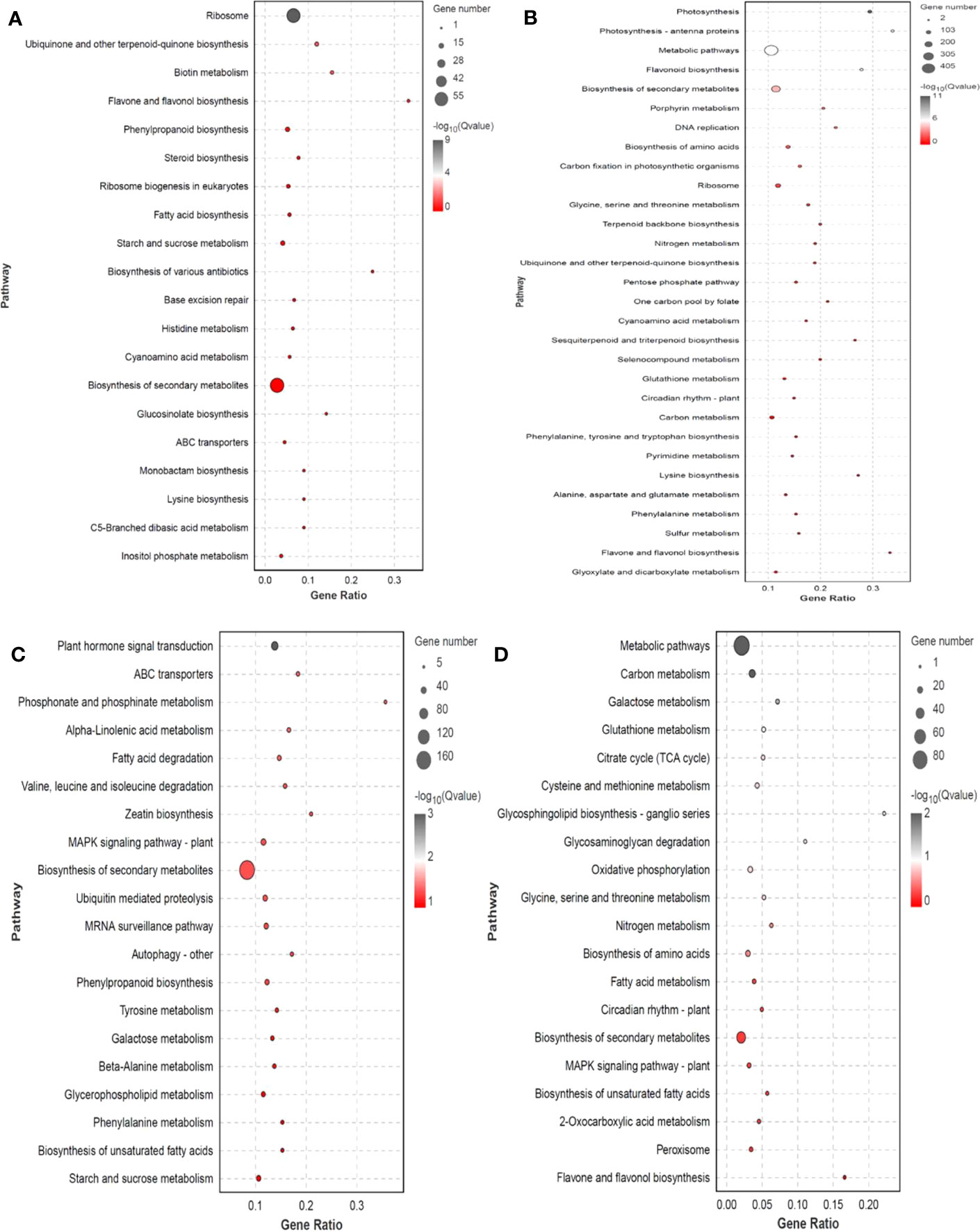
Figure 8 KEGG enrichment significance bubble chart of four trends (profiles 0, 1, 6, and 7) with significant differences after trend analysis. (A) profile 0 KEGG enrichment significance bubble charts; (B) profile 1 KEGG enrichment significance bubble charts; (C) profile 6 KEGG enrichment significance bubble charts; (D) profile 7 KEGG enrichment significance bubble charts.
Key mechanisms found under salt stress include the “glutathione metabolism,” and “plant hormone signal and MAPK signaling pathway - plant,”. The 14 genes encoding mitogen-activated protein kinases (MAPK), three genes encoding inhibitors of ethylene synthesis, one gene that indirectly affects the inhibition of phosphorylation in jasmonic acid synthesis (MYC2), 15 genes inhibiting dephosphorylation, and one gene indirectly inhibiting H2O2 synthesis (CAT1) in ABA synthesis under abiotic stress were upregulated. The “plant hormone signal transduction” pathway had 48 genes that were substantially enriched in Arabidopsis (P<0.05), four of which are involved in abscisic acid and ethylene, including those encoding protein phosphatase (PP2C), protein kinase (SnRK2), and ethylene receptors (ETR). Enzymes for the production and breakdown of auxin, cytokinin, brassinosteroid, jasmonic acid, and salicylic acid are encoded by other genes. A total of three genes are involved in GSSG reactions along the “glutathione metabolism” pathway, the three genes are related to GSH, one gene is involved in arginine biosynthesis, and one gene participates in glutamate metabolism. These genes were also upregulated (Supplementary Table 3; Figures 9A–C). In profile 0, DEGs were considerably enriched (P<0.05). Flavone and flavonol biosynthesis, as well as flavonoid biosynthesis, the expression levels of 25 DEGs encoding CHS, CHS1, CHS2, ANR, DRF, LAR, LDOX, F3H-2, CCOMT, and SALAT were downregulated (Supplementary Table 4; Figure 9D).
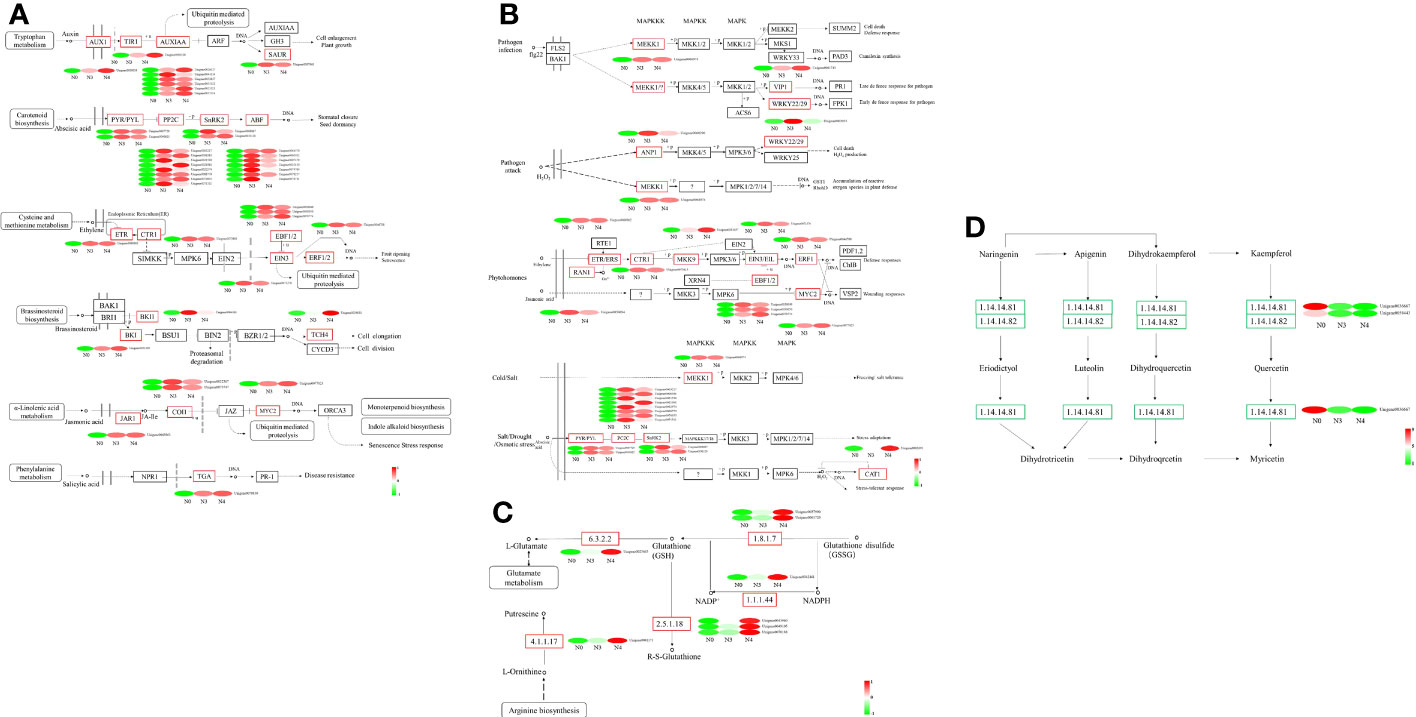
Figure 9 Heat map of differential gene expression in “the Plant hormone signal pathway”, “MAPK signaling pathway-plants”, “Glutathione metabolism pathway” and “flavone and flavonol biosynthesis pathway”. (A) the Plant hormone signal pathway, (B) MAPK signaling pathway-plants, (C) Glutathione metabolism pathway, (D) flavone and flavonol biosynthesis pathway. N0:0 mM/L NaCl stress; N3:600mM/L NaCl stress; N4:800mM/L NaCl stress.
RT-qPCR validates RNA-seq data
18 DEGs with diverse expression patterns were chosen for RT-qPCR to confirm the RNA-seq data (Supplementary Table 5). All of these potential genes were screened from N3 and N4, and qRT-PCR findings and RNA-seq data showed a good positive correlation (R2 = 0.871), indicating the RNA-seq data were repeatable and reliable (Figure 10).
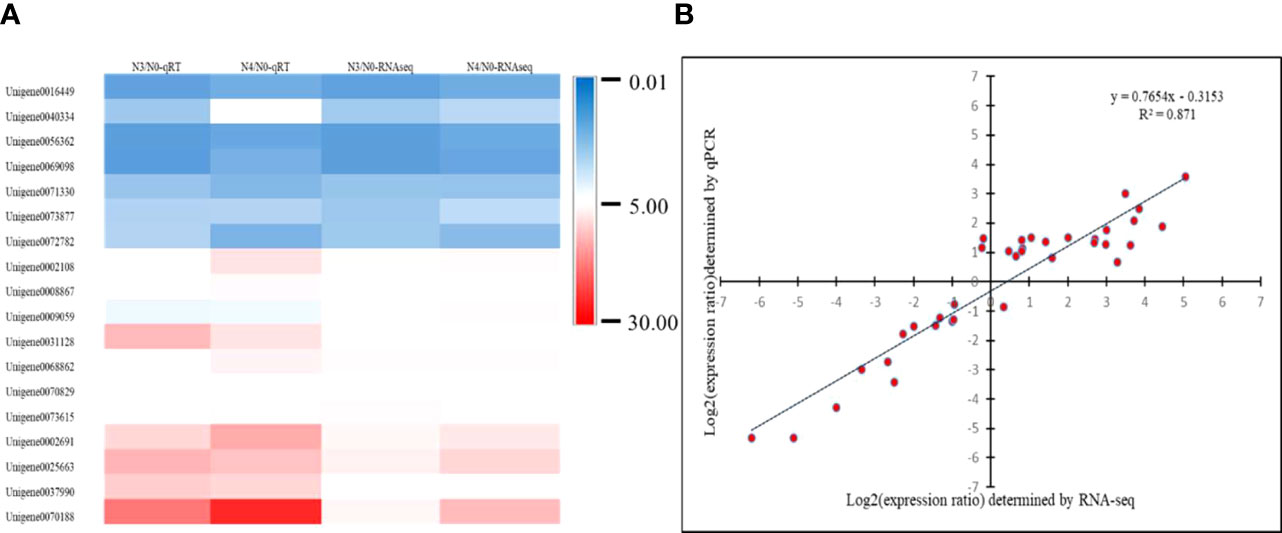
Figure 10 shows a heatmap of unigene expression to verify the accuracy of RT-qPCR. (A) Heatmap of 18 differently expressed unigenes’ expression levels; (B) correlation of 18 differentially expressed unigenes as determined by RNA sequencing and quantitative RT-qPCR. N0:0mM/L NaCl stress; N3:600mM/L NaCl stress; N4:800mM/L NaCl stress.
Discussion
Salt tolerance mechanisms of G. maritima
To adapt to an environment with high salt concentrations, some halophytes evolved specific salt-secreting structures (salt glands and salt vesicles). These salt-secreting structures secrete and store Na+ to protect the organisms from excessive Na+ toxicity (He et al., 2019). After crosscutting the leaves of G. maritima seedlings, salt gland tissue that secretes NaCl under salt stress was identified. However, the salt secretion mechanism of salt glands needs to be further explored. In the current work, we more thoroughly examined G. maritima’s physiological reaction to salt stress and predicted DEGs that are crucial for this organism’s ability to tolerate salt.
Research on the physiology of halophytes shows that the relative conductivity and proline content, which affect the stability of the plant membrane system, are important physiological indices for evaluating plant salt tolerance. Some studies have shown that changes in relative conductivity reflect the degree of damage to cell membranes under osmotic stress. The relative conductivities of Vaccinum corymbosum L., Graptoveria titubans, Ilex chinensis Sims., Xanthoceras sorbifolia Bunge., Miscanthus sinensis Anderss., and Triarrhena sacchariflora (Maxim.) Nakai leaves after salt stress (Wu, 2019; Zhang, 2019; Xie et al., 2020) have been measured, and the relative conductivity was found to tends to increase with increasing salt concentration, which is consistent with the measurement results of the relative conductivity of G. maritima leaves in the present study. This suggests that during salt stress, plant mesophyll cells amass a considerable amount of ions to maintain intracellular equilibrium and limit damage to the plant’s membranes. Under adverse conditions, plants accumulate excessive proline. Proline acts as an osmotic regulator to buffer salt, drought, and other stresses, or as a protector of cytoplasmic enzymes and membranes to minimize damage under adverse conditions. For example, with increasing NaCl concentration, the proline content in Pugionium cornutum (L.) Gaertn. leaves increases significantly (Yue et al., 2016). Rosmarinus officinalis leaf proline content increases dramatically with increasing stress concentration and time under NaCl (Chetouani et al., 2019). The proline content in Actinidia chinensis leaves show an upward trend compared to that in the control under high salt stress (Abid et al., 2020). We demonstrate here that G. maritima, when subjected to salt stress, has disruptions in its physiological and metabolic activity. The plant responded by changing its metabolite content. Proline accumulation was one such physiological response. When exposed to a stress concentration of more than 600 mM NaCl, however, the cellular balance of osmotic control and ion regulation was disrupted. the metabolism in cells was disordered, and the proline content was reduced, indicating that the stability of enzymes and the membranes of plant cells may have been lost, and the plant itself could not grow normally at this concentration.
Plants boost antioxidant enzyme activity, eliminate excess ROS, and keep the plant’s internal dynamic equilibrium stable while under stress to prevent cell membrane damage. In order to get rid of unwanted ROS and adapt to adversity, plants rely on antioxidant enzyme systems including SOD, POD, and CAT. Some studies have shown that, as the NaCl stress concentration increases, SOD activity in plant leaves first increases and then decreases. Evidence like this demonstrates that plants under salt stress first respond by keeping their SOD activity high in order to prevent harm; however, high concentrations (800 mM) of salt stress produce O2- exceeding the capacity of plants to compensate, dynamic balance is destroyed, and SOD activity is reduced (Zhu et al., 2021). This is consistent with the observed changes in SOD activity in the leaves of G. maritima in the present study. As an antioxidant enzyme that further scavenges the antioxidant products of SOD in cells, POD activity first increases and then decreases with increasing salt concentrations. High POD activity was maintained in the leaves of G. maritima seedlings exposed to salt stress, and H2O2 was cleared in time to prevent more significant OH- toxicity. The normal development of G. maritima seedlings was disrupted by exposure to high quantities because of a reduction in POD activity in the leaves, a loss in the plants’ capacity to withstand ROS. The same has also been reported for POD activity changes in plants, such as Dalbergia granadillo, Poa pratensis L., and Hibiscus syriacus L. under salt damage (Zhao et al., 2017; Lai et al., 2020). CAT is an enzyme scavenger that can remove H2O2 in plants, promote its decomposition into molecular oxygen and H2O, and reduce the toxic effects of H2O2 on plant cells. Under salt stress, CAT activity in the leaves of Suaeda glauca (Bunge) Bunge., Matthiolaincana L. R.Br., Hibiscus syriacus L., and Mentha haplocalyx B. first increases and then decreases, which is generally consistent with our results (Jafari and Hashemi Garmdareh, 2019, Pang et al., 2005, Haddadi et al., 2016; Li et al., 2020). This illustrates that when salt concentration grows, the antioxidant system initially acts properly to promote CAT activity, and later, under high salt stress, it can no longer operate normally due to increased H2O2, resulting in decreased CAT activity, which is similar to the findings of Hibiscus syriacus L. (Lu, 2021).
For this research, we used high-throughput transcriptome sequencing to investigate the processes through which G. maritima is able to withstand high concentrations of salt. Genes possibly involved in salt stress were identified using the anatomy, physiology, and biochemistry of G. maritima leaves as a reference (Zhao et al., 2005). Through the analysis of physiological and biochemical indexes, proline content and SOD and CAT activities in the leaves of G. maritima under salt stress were found to be significantly different from those in the control. Transcriptome analysis shows that there were 11 genes affecting proline synthesis, SOD, and CAT activity in the “arginine and proline metabolism” pathway and “peroxisome” pathway, among which six genes affected proline content and regulated SOD and CAT activity. POX2 expression is consistent with earlier studies indicating that under stress, plants upregulate genes involved in proline synthesis while downregulating those involved in degradation. This gene mainly negatively regulates the activity of proline dehydrogenase (PRODH) to inhibit the degradation of proline to P5C and maintain a high level of proline content to resist the damage caused by salt. In the process of proline production by glutamic acid, the four genes regulating P5CS and GSA were closely related to increased proline content in G. maritima after salt stress. Three PRO2 genes were upregulated, and one gene was downregulated. P5CS regulatory genes that affect changes in proline content (such as AtP5CS1, OsP5CS, MsP5CS-1, MsP5CS-2, OmPro1, TomPro2, SsP5CS, MtP5CS1, MtP5CS2, PvP5CS1, and PvP5CS2) have also been found in Arabidopsis thaliana, Oryza sativa L., Lycopersicon esculentum, Suaeda salsa (L.) Pall., diploid alfalfa, and Phaseolus vulgaris L. After salt stress, gene expression and proline content in plants are significantly increased compared to those in the control. In the ornithine pathway, salt stress promotes δ-Oat gene expression to increase the conversion of ornithine aminotransferase to GSA, similar to the findings of Nancy (Roosens et al., 1998). A study of δ-ornithine aminotransferase in Arabidopsis under salt stress show similar results. In the present study, the five genes affecting the antioxidant enzyme system included two upregulated catalases (CAT1 and CAT3), two upregulated superoxide dismutases (SOD1 (Gu/Zn SOD), and SOD2 (Fe/Mn SOD)), and one downregulated superoxide dismutase (SOD1 (Gu/Zn SOD)). SOD is a metal enzyme. Salt stress induced the expression of Gu/Zn SOD and Fe/Mn SOD genes, which is similar to the expression of antioxidant enzyme genes in Lycopersicon esculentum and Vigna radiata (Guan et al., 2017; Zhu et al., 2021). CAT is an important antioxidant enzyme system. For example, in rice, the expression of CATA and CATC is controlled by circadian rhythm. Under salt stress, CATC regulates H2O2 homeostasis and improves salt tolerance in rice (Tounsi et al., 2019). In Chenopodium quinoa willd., 4 genes of the CAT family are upregulated to deal with the damage caused by salt stress. This coincides with the findings from the current investigation on the expression of the two genes known to influence CAT activity. The above results show that some important biological activities are related to the transcriptional expression of proline, SOD, and CAT genes that regulate plant salt tolerance (Sun et al., 2021).
Main genes affecting salt tolerance of G. maritima seedlings
In addition to participating in the antioxidant process, G. maritima’s tolerance to salt is influenced by changes in the amount of the plant hormone ABA and the MAPK signaling pathway. Responses to salt stress are regulated in part by plant hormones (Ryu and Cho, 2015) A survey of genes involved in hormone signaling pathways has shown that they have a role in G. maritima’s salt tolerance. Several plant species have had their salt tolerance increased by evidence showing that ABA and JA were present (Goossens et al., 2016; Vishwakarma et al., 2017; Yang et al., 2017). In plants, ABA plays a crucial role as a regulator of signal transduction processes (Sicilia et al., 2019). As a result of stimulus, ABA-bound PYR/PYL/RCAR inhibits PP2Cs, activating SnRK2. As a double-negative regulator, this protein complex raises the expression of linked genes in response to ABA (Huang et al., 2018). NaCl stress was shown to cause an increase in the expression of the ABF gene in G. maritima via downregulating the activity of SnRK2. Previous studies on plants have shown the same results, thus these results are reliable (Yang et al., 2017; Lei et al., 2018; Lei et al., 2021). Numerous studies have revealed that exogenous ABA administration in plants activates the MAPK pathway, which is then responsible for transcriptional regulation. This suggests that the MAPK pathway plays an important role in ABA signal transduction (Zhang et al., 2014; Li et al., 2016; Li et al., 2017). The MEK1-MKK2-MPK6/MPK4 pathway improves salt tolerance in Arabidopsis via an interaction between MKK2 and MPK4 (Teige et al., 2004). The MAPK cascade MEKK1-MKK2-MPK6/MPK4 plays a critical role in regulating ROS balance and stress signal transmission during salt stress by responding to SA and rosin (Yang and Guo, 2018a). After 4 hours of exposure to salt stress, cotton’s MAPK cascade gene expression is considerably upregulated (Chen et al., 2020). The initial step in ABA signal transduction involves the PYR/PYL/RCAR protein and phosphatase/kinase pairs PP2Cs and SnRK2s, respectively, with antagonistic functions. ABA signal transduction results in transcription factors activating gene expression under the control of SnRK2s (Cutler et al., 2010; Finkelstein, 2013; Tan et al., 2018). In the present study, after G. maritima was subjected to salt stress, PP2C was upregulated and inhibited the activity of SnRK2 kinase. The expression of MAPKK17-18 in the downstream MAPK signaling pathway was promoted, alleviating leaf senescence and enhancing the salt tolerance of G. maritima. This is similar to the findings of a previous study on Arabidopsis (Matsuoka et al., 2015).
In addition to regulating the SOD and CAT processes, glutathione metabolism is an important regulator. Oxidized glutathione (GSSG) is converted back to glutathione by GR catalysis (GSH) (Tao et al., 2003). Neto showed that GR activity in plant leaves increases significantly after salt stress treatment compared with that in the control (De et al., 2006). Huang found that tomato seedlings under 80 mM salt treatment resist salt stress by significantly enhancing the activities of GR, MDHAR, APX, and DHAR in chloroplasts (Huang and Shan, 2018). Under high salt stress, an increase in plant ROS leads to cell membrane lipid peroxidation. The level of MDA indicates the extent to which plant membranes have been damaged by salt stress. MDA is the primary byproduct of membrane lipid peroxidation. Valentina discovered that in salt-tolerant tomato plants, GSH production and accumulation are triggered by salt stress. Additionally, significant amounts of GSH, GPX, GR, and GSSG are present in salt-tolerant tomato, which helps to reduce membrane lipid peroxidation due to salt stress (Mittova et al., 2003). Analysis of transcriptome data showed that modulation of eight genes involved in glutathione metabolism was a major mechanism impacting G. maritima’s salt tolerance. These genes are involved in regulating the activity of GR and the synthesis of GSH. After being put through the ringer by biological and abiotic stresses, plants create flavonoids and flavonoids to eliminate H2O2, boost plant antioxidant activity, minimize water loss, improve the environment in plant cells, and resist damage, as proven in several studies (Nakabayashi et al., 2014). However, by transcriptome analysis of G. maritima, the three genes involved in flavonoid, flavone, and flavonol biosynthesis, 4,5,6,7-trihydroxyflavonol, quercetin, and myricetin, were found to be downregulated. A study also showed that sea milkweed does not improve its antioxidant activity through flavonoids after salt stress.
Conclusion
In this study, G. maritima was a salt secreting plant, and the salt secreting tissue in its leaves was salt glands, which sank into epidermal cells and consisted of stem cells, collecting cells, and secreting cells. After high salt stress, the leaf cells of G. maritima grass show a decrease in volume and become densely arranged. An increase in salt concentration will increase the water loss effect of the leaves. After high salt stress, a large amount of proline is produced in the leaves to regulate osmotic balance, and the activities of SOD and POD enzymes in the antioxidant enzyme system are significantly increased to eliminate the large amount of ROS generated due to salt damage, ensuring normal growth in a high salt environment. A study on the molecular mechanism of sea milkweed leaves revealed 11 DEGs that regulate proline synthesis and CAT and SOD activities, which may be key genes affecting salt tolerance of G. maritima. It is also located in Plant hormone signaling, Glutathione metabolism and MAPK signaling pathway-plant. The KEGG pathway, including flavonoid biosynthesis, flavonoid biosynthesis, and flavonol biosynthesis, may also be important metabolic pathways that affect salt tolerance of G. maritima. Studying and analyzing 93 DEGs in these pathways can lay the foundation for further exploring salt tolerance genes and improving salt tolerance in forage crops. It also provides precious biological improvement resources for improving Alkali soil.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
FS designed the experiment, RG, ZW, XL and YY performed the experiments, RG and FT analyzed the data, RG wrote the manuscript. All authors reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Key Projects in Science and Technology of Inner Mongolia (2021ZD0031) and the Science and Technology Project of the Inner Mongolia Autonomous Region (2020GG0063).
Acknowledgments
Thanks to the Key Laboratory of Grassland Resources, Ministry of Education of P.R. of China for providing facilities. The authors would like to thank Editage (www.editage.cn) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1173191/full#supplementary-material
References
Abid, M., Zhang, Y. J., Li, Z., Bai, D. F., Fang, J. B. (2020). Effect of Salt stress on growth, physiological and biochemical characters of Four kiwifruit genotypes. Scientia Horticulturae. 271, 109473. doi: 10.1016/j.scienta.2020.109473
Beers, R. F., Sizer, I. W. (1952). A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. Biol. Chem. 195, 133–140. doi: 10.1016/S0021-9258(19)50881-X
Calone, R., Bregaglio, S., Sanoubar, R., Noli, E., Barbanti, L. (2021). Physiological adaptation to water salinity in six wild halophytes suitable for mediterranean agriculture. Plants 10 (2), 309. doi: 10.3390/plants10020309
Chance, B., Maehly, A. C. (1955). Assay of catalases and peroxidases. Methods Enzymol. 2, 764–775. doi: 10.1016/S0076-6879(55)02300-8
Chen, L., Sun, H., Wang, F., Yue, D., Yang, X. (2020). Genome-wide identification of MAPK cascade genes reveals the GhMAP3K14–GhMKK11–GhMPK31 pathway is involved in the drought response in cotton. Plant Mol. Biol. 103 (9), 211–223. doi: 10.1007/s11103-020-00986-0
Chetouani, M., Mzabri, I., Aamar, A., Boukroute, A., Kouddane, N., Berrichi, A. (2019). Morphological-physiological and biochemical responses of Rosemary (Rosmarinus officinalis) to salt stress. Materials Today: Proc. 13, 752–761. doi: 10.1016/j.matpr.2019.04.037
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., Abrams, S. R. (2010). Abscisic acid, emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
De, A. N. A. D., Prisco, J. T., Eneas-Filho, J., Abreu, C. E., Gomes-Filho, E. (2006). Effect of salt stress on antioxidative enzymes and lipid peroxidation in leaves and roots of salt-tolerant and salt-sensitive maize genotypes. Environ. Exp. Botany. 56 (1), 87–94. doi: 10.1016/j.envexpbot.2005.01.008
Du, C., Zhang, J., Zhang, F. C. (2017). Expression analysis of Rev1 and Rev3 of Halostachys caspica under salt stress. Bull. Botanical Res. 37 (2), 211–215,226. doi: 10.7525./j.issn.1673-5102.2017.02.008
Finkelstein, R. (2013). Abscisic Acid synthesis and response. Arabidopsis book 11, e0166. doi: 10.1199/tab.0166
Ghanem, E., Mohamed, E., Kasem, A., El-Ghamery, A. A. (2021). Differential salt tolerance strategies in three halophytes from the same ecological habitat: augmentation of antioxidant enzymes and compounds. Plants 10 (6), 1100. doi: 10.3390/plants10061100
Gong, B., Li, X., Bloszies, S., Wen, D., Sun, S. S., Wei, M., et al. (2014). Sodic alkaline stress mitigation by interaction of nitric oxide and polyamines involves antioxidants and physiological strategies in Solanum lycopersicum. Free Radic. Biol. Med. 71, 36–48. doi: 10.1016/j.freeradbiomed.2014.02.018
Goossens, J., Fernández-Calvo, P., Schweizer, F., Goossens, A. (2016). Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 91 (6), 673–689. doi: 10.1007/s11103-016-0480-9
Guan, Q., Liao, X., He, M., Li, X., Wang, Z., Ma, H., et al. (2017). Tolerance analysis of chloroplast OsCu/Zn-SOD overexpressing rice under NaCl and NaHCO3 stress. PloS One 12 (10), e0186052. doi: 10.1371/journal.pone.0186052
Haddadi, B. S., Hassanpour, H., Niknam, V. (2016). Effect of salinity and waterlogging on growth, anatomical and antioxidative responses in Mentha aquatica L. Acta Physiologiae Plantarum 38 (5), 1–11. doi: 10.1007/s11738-016-2137-3
He, F. L., Bao, A. K., Wang, S. M. (2019). NaCl stimulates growth and alleviates drought stress in the salt-secreting xerophyte Reaumuria soongorica. Environ. Exp. Bot. 162, 433–443. doi: 10.1016/j.envexpbot.2019.03.014
Huang, K. C., Lin, W. C., Cheng, W. H. (2018). Salt hypersensitive mutant 9, a nucleolar APUM23 protein, is essential for salt sensitivity in association with the ABA signaling pathway in Arabidopsis. BMC Plant Biol. 18 (1), 40. doi: 10.1186/s12870-018-1255-z
Huang, G., Shan, C. (2018). Lanthanum improves the antioxidant capacity in chloroplast of tomato seedlings through ascorbate-glutathione cycle under salt stress. Scientia Horti culturae. 232, 264–268. doi: 10.1016/j.scienta.2018.01.025
Jafari, S., Hashemi Garmdareh, S. E. (2019). Effects of salinity on morpho-physiological, and biochemical characteristics of stock plant (Matthiola incana L.). Scientia Hortic. 257 (11), 108731. doi: 10.1016/j.scienta.2019.108731
Jin, H., Dong, D., Yang, Q., Zhu, D. (2016). Salt-responsive transcriptome profiling of suaeda glauca via rna sequencing. PloS One 11 (3), e0150504. doi: 10.1371/journal.pone.0150504
Khan, M. K., Pandey, A., Hamurcu, M., Avsaroglu, Z. Z., Ozbek, M., Omay, A. H., et al. (2021). Variability in physiological traits reveals boron toxicity tolerance in aegilops species. Front. Plant Sci. 12, 736614. doi: 10.3389/fpls.2021.736614
Kuang, L., Shen, Q., Wu, L., Yu, J., Fu, L., Wu, D., et al. (2019). Identification of microRNAs responding to salt stress in barley by high-throughput sequencing and degradome analysis. Environ. Exp. Bot. 160, 59–70. doi: 10.1016/j.envexpbot.2019.01.006
Lai, X. L., Yan, Y. J., Yan, L. H. (2020). Effects of NaCl stress on growth and physiological characteristics of Dalbergia hupeana seedlings. Plant Physiol. 56 (2), 309–316. doi: 10.13592/j.cnki.ppj.2019.0152
Lei, P., Liu, Z., Hu, Y., Kim, H. C., Meng, F. (2021). Transcriptome analysis of salt stress responsiveness in the seedlings of wild and Cultivated Ricinus communis L. Biotechnol. 327, 106–116. doi: 10.1016/j.jbiotec.2020.12.020
Lei, Y., Xu, Y., Hettenhausen, C., Lu, C., Shen, G., Zhang, C. (2018). Comparative analysis of alfalfa (Medicago sativa L.) leaf transcriptomes reveals genotype-specific salt tolerance mechanisms. BMC Plant Biol. 18 (1), 35. doi: 10.1186/s12870-018-1250-4
Li, Q., Wang, Y., Liu, C., Pei, Z. M., Shi, W. L. (2017). The crosstalk between ABA, nitric oxide, hydrogen peroxide, and calcium in stomatal closing of Arabidopsis thaliana. Biologia 72 (10), 1140–1146. doi: 10.1515/biolog-2017-0126
Li, Z., Xu, C., Wang, J. (2020). Integrated physiological, transcriptomic and proteomic analyses revealed molecular mechanism for salt resistance in Solidago canadensis L. Environ. Exp. Botany. 179 (6), 104211. doi: 10.1016/j.envexpbot.2020.104211
Li, K., Yang, F., Zhang, G., Song, S., Li, Y., Miao, Y., et al. (2016). AIK1, A mitogen-activated protein Kinase, Modulates Abscisic Acid Responses through the MKK5-MPK6 kinase Cascade. Plant Physiol. 173, 1391–1408. doi: 10.1104/pp.16.01386
Liu, P. (2006). Study on Adptational Anatomical Structure of Nutritive Organs and the Salt-tolerance Physiology in Two Species of Limonium (Xinjiang, China: Shihezi University).
Lu, W. J. (2021). Effects of NaCl stress on physiological and biochemical characteristics of two Hibiscus cultivars (Shandong, China: Shandong Agricultural University). Master’s degree thesis.
Ma, X., Li, W., Mao, W., Peng, H. (2003). Difference in relative conductivity and ultrastructure of leaf between two wheat cultivars with different thermotolerance under heat acclimation and heat stress. China Agric. Univ. 8 (5), 4–8. doi: 1007-4333(2003)05-0004-05
Matsuoka, D., Yasufuku, T., Furuya, T., Nanmori, T. (2015). An abscisic acid inducible arabidopsis mapkkk, mapkkk18 regulates leaf senescence via its kinase activity. Plant Mol. Biol. 87 (6), 565–575. doi: 10.1007/s11103-015-0295-0
McCord, J. M., Fridovich, I. (1969). Superoxide dismutase an enzymic function for erythrocuprein (hemocuprein). Biol. Chem. 244, 6049–6055. doi: 10.1016/S0021-9258(18)63504-5
Mittova, V., Theodoulou, F. L., Kiddle, G., Gómez, L., Volokitaa, M., Talc, M., et al. (2003). Coordinate induction of glutathione biosynthesis and glutathione-metabolizing enzymes is correlated with salt tolerance in tomato. FEBS Letters. 554 (3), 417–421. doi: 10.1016/S0014-5793(03)01214-6
Munns, R., Day, D. A., Fricke, W., Watt, M., Arsova, B., Barkla, B. J. (2019). Energy costs of salt tolerance in crop plants. New Phytol. 225, 1072–1090. doi: 10.1111/nph.15864
Nakabayashi, R., Yonekura-Sakakibara, K., Urano, K., Suzuki, M., Yamada, Y., Nishizawa, T., et al. (2014). Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77 (3), 367–379. doi: 10.1111/tpj.12388
Nakano, Y., Asada, K. (1981). Hydrogen peroxide is scavenged by ascorbatespecific peroxidase in spinach chloroplasts. Plant Cell Physiol. 22, 867–880. doi: 10.1093/oxfordjournals.pcp.a076232
Pandey, A., Khan, M. K., Hamurcu, M., Brestic, M., Topal, A., Gezgin, S. (2022). Insight into the Root Transcriptome of a Boron-Tolerant Triticum zhukovskyi Genotype Grownunder Boron Toxicity. Agronomy 12, 2421. doi: 10.3390/agronomy12102421
Pang, C. H., Zhang, S. J., Gong, Z. Z., Wang, B. S. (2005). NaCl treatment markedly enhances H2O2-scavenging system in leaves of halophyte Suaeda salsa. Physiologia Plantarum 125 (4), 490–499. doi: 10.1111/j.1399-3054.2005.00585.x
Rebhi, A. E. M., Lounici, H., Lahrech, M. B., Morel, J. L. (2019). Response of Artemisia herba alba to hexavalent chromium pollution underarid and semi-arid conditions. Int. J. Phytoremediation 21, 224–229. doi: 10.1080/15226514.2018.1524841
Roosens, N. H. C. J., Thu, T. T., Jacobs, I. M. (1998). Isolation of the ornithine-δ-aminotransferase cdna and effect of salt stress on its expression in Arabidopsis thaliana. Plant Physiol. 117 (1), 263–271. doi: 10.1104/pp.117.1.263
Rozema, J. (1975). Eco-physiological investigation into the salt tolerance of Glaux maritima L. Plant Biol. 24 (5-6), 407–416. doi: 10.1111/j.1438-8677.1975.tb01031.x
Rozema, J., Sminia, R. T. (1977). A light and electron-microscopical study on the structure and function of the salt gland of Glaux maritima L. New Phytol. 79 (3), 665–671. doi: 10.1111/j.1469-8137.1977.tb02251.x
Ryu, H., Cho, Y. G. (2015). Plant hormones in salt stress tolerance. Plant Biol. 58 (3), 147–155. doi: 10.1007/s12374-015-0103-z
Saddhe, A. A., Manuka, R., Nikalje, G. C., Penna, S. (2020). Halophytes as a potential resource for phytodesalination. Handb. Halophytes., 1–21. doi: 10.1007/978-3-030-17854-3_92-1
Safdar, H., Amin, A., Shafiq, Y., Ali, A., Yasin, R., Sarwar, M. I., et al. (2019). A review: impact of salinity on plant growth. Nat. Sci. 1, 34–40. doi: 10.7537/marsnsj170119.06
Shabala, S., Bose, J., Hedrich, R. (2014). Salt bladders: do they matter? Trends Plant Sci. 19, 687–691. doi: 10.1016/j.tplants.2014.09.001
Shrivastava, P., Kumar, R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Sicilia, A., Testa, G., Santoro, D. F., Cosentino, S. L., Lo Piero, A. R. (2019). RNA-Seq analysis of giant cane reveals the leaf transcriptome dynamics under long-term salt stress. BMC Plant Biol. 19 (1), 355. doi: 10.1186/s12870-019-1964-y
Sun, M., Han, J. W., Wang, M. L., Zhang, L. P., Zhang, X., Guo, S. L., et al. (2021). Identification of CAT gene family in chenopodium quinoa. Mol. Plant Breed. 19 (18), 5933–5942. doi: 10.13271/j.mpb.019.005933
Tan, W., Zhang, D., Zhou, H., Zheng, T., Yin, Y., Lin, H. (2018). Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PloS Genet. 14 (4), e1007336. doi: 10.1371/journal.pgen.1007336
Tao, J., Li, T., Sun, C. B., Zhong, C. Z., Qin, C. Y., Chen, S. G., et al. (2003). Review on salt stress in plant. Journal of Jilin Forestry Science and Technology 32 (5), 1–7. doi: 10.16115/j.cnki.issn.100–129.2003.05.001
Teige, M., Scheikl, E., Eulgem, T., Dóczi, R., Hirt, H. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15 (1), 141–152. doi: 10.1016/j.molcel.2004.06.023
Tlili, A., Ghanmi, E., Ayeb, N., Louhaichi, M., Tarhouni, M. (2020). Revegetation of marginal saline rangelands of southern Tunisia using pastoral halophytes. Afr. J. Range Forage Sci. 37 (2), 151–157. doi: 10.2989/10220119.2020.1720293
Tounsi, S., Kamoun, Y., Feki, K., Jemli, S., Say di, M. N., Ziadi, H., et al. (2019). Localization and expression analysis of a novel catalase from Triticum monococcum TmCAT1 involved in response to different environmental stresses. Plant Physiol. Biochem. 139, 366–378. doi: 10.1016/j.plaphy.2019.03.039
Trofimova, T. A., Hossain, A., Silva, J. (2012). The ability of medical halophytes to phytoremediate soil contaminated by salt and heavy metals in Lower Volga, Russia. Plant Sci. Biotechnol. 6, 108–114.
Vishwakarma, K., Upadhyay, N., Kumar, N., Yadav, G., Singh, J., Mishra, R. K., et al. (2017). Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8, 161. doi: 10.3389/fpls.2017.00161
Walker, D. J., Lutts, S., Sánchezgarcía, M., Correal, E. (2014). Atriplex halimus l.: its biology and uses. Arid Environments 100-101 (1), 111–121. doi: 10.1016/j.jaridenv.2013.09.004
Wilson, H., Mycock, D., Weiersbye, I. M. (2017). The salt glands of Tamarix usneoides E. Mey. ex Bunge (South African salt cedar). Int. J. Phytoremediat 19, 587–595. doi: 10.1080/15226514.2016.1244163
Wu, F. Z. (2019). Growth and physiological response and salt-tolerance differences of three blueberry seedlings under salt stress. Plant Physiol. J. 55 (11), 1638–1646. doi: 10.13592/j.cnki.ppj.2019.0301
Wu, X. X., Bai, T. H., Zhang, L., Bao, A. K. (2020). Research progress on mechanisms underlying salt secretion in recretohalophytes. Plant Physiol. J. 56 (12), 2526–25325. doi: 10.13592/j.cnki.ppj.2020.0370
Xie, X. Q., Lin, Q. J., Wang, M. S., Chen, H. D., Sun, J. Q. (2020). Physiological responses of six foreign Wild Peony plants to NaCl stress. Fujian Sci. Technol. Trop. Crops 45 (2), 26–28. doi: 1006-2327-(2020)02-0026-03
Xie, Q., Niu, J., Xu, X. L., Xu, L. X., Zhang, Y. B., Fan, B. (2015). De novo assembly of the Japanese lawngrass (Zoysia japonica Steud.) root transcriptomeand identification of candidate unigenes related to early responses under salt stress. Front. Plant Science. 6, 610. doi: 10.3389/fpls.2015.00610
Yamamoto, K., Matsutani, M., Shiwa, Y., Ishige, T., Sakamoto, H., Saitoh, H., et al. (2020). Comparative Analysis of Bacterial Diversity and Community Structure in the Rhizosphere and Root Endosphere of Two Halophytes, Salicornia europaea and Glaux maritima, Collected from Two Brackish Lakes in Japan. Microbes Environ. 35 (3), ME20072. doi: 10.1264/jsme2.ME20072
Yamamoto, K., Shiwa, Y., Ishige, T., Sakamoto, H., Tanaka, K., Uchino, M., et al. (2021). Bacterial diversity associated with the rhizosphere and endosphere of two halophytes: Glaux maritima and Salicornia europaea. Front. Microbiol. 9, 2878. doi: 10.3389/fmicb.2018.02878
Yang, Y. Q., Guo, Y. (2018a). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217 (2), 523–539. doi: 10.1111/nph.14920
Yang, Z., Lu, R., Dai, Z., Yan, A., Tang, Q., Cheng, C., et al. (2017). Salt-stress response mechanisms using de novo transcriptome sequencing of salt-tolerant and sensitive Corchorus spp. Genotypes. Genes (Basel). 8 (9), 226. doi: 10.3390/genes8090226
Yuan, Y., Chen, M., Chen, Q. (2012). Effects of nacl stress on physiological and biochemical characteristics of pitaya (hylocereus undatus) seedlings. Chin. Trop. Crops. 33(07), 1256–1260. doi: 10.3969/j.issn.1000-2561.2012.07.021
Yuan, F., Yu, L., Leng, B. Y., Zheng, G. Y., Feng, Z. T., Li, P. H., et al. (2015). Comparative transcriptome analysis of developmental stages of the limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 38 (8), 1637–1657. doi: 10.1111/pce.12514
Yue, L. J., Cui, Y. N., Yuan, K., Kang, J. J., Wang, S. M. (2016). The osmotic adjustment in pugionium cornutum subjected to salt stress. Plant Physiol. J. 52 (4), 569–574. doi: 10.13592/j.cnki.ppj.2016.0028
Zhang, J. (2019). Effects of saline-alkali stress on growth physiology of eight ilex species. (dissertation) (Changsha, China: Central south university of forestry and technology, Changsha, China).
Zhang, J., Zou, D., Li, Y., Sun, X., Wang, N. N., Gong, S. Y., et al. (2014). GhMPK17, a cotton mitogen-activated protein kinase, is involved in plant response to high salinity and osmotic stresses and ABA signaling. PloS One 9, e95642. doi: 10.1371/journal.pone.0095642
Zhao, S. Y. (2016). The study of salt-secreting mechanisms underlying Reaumuria soongorica in response to salt and drought (dissertation) (Lanzhou: Lanzhou University).
Zhao, L. Y., Deng, X. P., Shan, L. (2005). The response mechanism of active oxygen species removing system to drought stress. Acta Bot. Boreal-Occident. Sin. 25 (2), 413–418. doi: 1000-4025.2005.02-0413-06
Zhao, H., Jia, F., Zhang, F., Wang, Y. (2014). The transcriptome information analysis of differentially expressed genes of halostachys caspica under salt stress. Chin. J. Bioinf. 12 (02), 90–98. doi: 10.3969/j.issn.1672-5565.2014.02.03
Zhao, H. Y., Wang, J. S., Liu, L. Q., Chen, Y. Q., Mengfei, L. I., Quanwei, L. U. (2017). Morphological and physiological mechanism of salt tolerance in gossypium barbadense to salt stress at seedling stage. Scientia Agricultura Sinica. 50 (18), 3494–3505. doi: 10.3864/j.issn.0578-1752.2017.18.006
Zhou, S., Han, J. L., Zhao, K. F. (2001). Advance of study on recretohalophytes. Chin. J. Appl. Environ. Biol. 7, 496–501. doi: 10.3321/j.issn:1006-687X.2001.05.020
Zhu, X. T., Huang, Y., Lu, Y., He, D. M., Li, H. X., Gu, X. R., et al. (2021). Bioinformatics analysis of SOD gene in mung bean and its expression under salt stress. Mol. Plant Breed. 21 (15), 4886–4894. doi: 10.13271/j.mpb.021.004886
Keywords: Glaux maritima, salt gland, physiology, transcriptome analysis, DEGs
Citation: Gu R, Wan ZQ, Tang F, Liu XT, Yang Yt and Shi Fl (2023) Physiological and transcriptomic analysis of salt tolerant Glaux maritima grown under high saline condition. Front. Plant Sci. 14:1173191. doi: 10.3389/fpls.2023.1173191
Received: 24 February 2023; Accepted: 14 August 2023;
Published: 29 August 2023.
Edited by:
Supratim Basu, New Mexico Consortium, United StatesReviewed by:
Mariana Reginato, National University of Río Cuarto, ArgentinaMohd. Kamran Khan, Selçuk University, Türkiye
Copyright © 2023 Gu, Wan, Tang, Liu, Yang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng ling Shi, nmczysfl@126.com
 Rui Gu
Rui Gu Zhi Qiang Wan
Zhi Qiang Wan Fang Tang1
Fang Tang1