Abstract
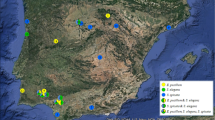
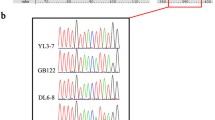
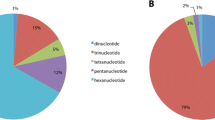
Plasmodial slime molds are members of the class Amoebozoa forming elaborate fruit bodies releasing airborne spores. Two species concepts have been developed independently: a morphological relying on fruit body characters, and a biological relying on crossing studies of a few cultivable species. In an attempt to reconcile both concepts, we obtained for 198 specimens of the common species Trichia varia partial sequences of three independent markers (nuclear small-subunit (SSU) ribosomal RNA gene, extrachromosomal; elongation factor 1 alpha gene, chromosomal; cytochrome oxidase subunit 1 gene, mitochondrial). The resulting phylogeny revealed 21 three-marker genotypes clustering into three groups. Combinations of the single-marker genotypes occurred exclusively within these groups, called 1, 2a, and 2b. To examine the suitability of group I introns to monitor speciation events, complete SSU sequences were generated for 66 specimens, which revealed six positions that can carry group I introns. For each of the groups 1 and 2a, five of these positions were occupied by different intron genotypes; and no genotype was shared by the two groups. Group 2b was devoid of introns. Putatively functional or degenerated homing endonuclease genes were found at different positions in groups 1 and 2a. All observations (genotypic combinations of the three markers, signs of recombination, intron patterns) fit well into a pattern of three cryptic biological species that reproduce predominantly sexual but are reproductively isolated. The pattern of group I introns and inserted homing endonuclease genes mounts evidence that the Goddard-Burt intron life cycle model applies to naturally occurring myxomycete populations.





Similar content being viewed by others
References
Adl, S. M., Simpson, A. G., Lane, C. E., Lukeš, J., Bass, D., Bowser, S. S., et al. (2012). The revised classification of eukaryotes. Journal of Eukaryotic Microbiology, 59, 429–493.
Agapow, P. M., & Burt, A. (2001). Indices of multilocus linkage disequilibrium. Molecular Ecology Notes, 1, 101–102.
Aguilar, M., Fiore-Donno, A. M., Lado, C., & Cavalier-Smith, T. (2013). Using environmental niche models to test the ‘everything is everywhere’ hypothesis for Badhamia. The ISME Journal, 8, 737–745.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402.
Betterley, D. A., & Collins, O. N. R. (1983). Reproductive systems, morphology, and genetical diversity in Didymium iridis (Myxomycetes). Mycologia, 75, 1044–1063.
Bhattacharya, D., Friedl, T., & Damberger, S. (1996). Nuclear-encoded rDNA group I introns: origin and phylogenetic relationships of insertion site lineages in the green algae. Molecular Biology and Evolution, 13, 978–989.
Bhattacharya, D., Cannone, J. J., & Gutell, R. R. (2001). Group I intron lateral transfer between red and brown algal ribosomal RNA. Current Genetics, 40, 82–90.
Bhattacharya, D., Reeb, V., Simon, D. M., & Lutzoni, F. (2005). Phylogenetic analyses suggest reverse splicing spread of group I introns in fungal ribosomal DNA. BMC Evolutionary Biology, 5, 68.
Brown, M. W., Silberman, J. D., & Spiegel, F. W. (2012). A contemporary evaluation of the acrasids (Acrasidae, Heterolobosea, Excavata). European Journal of Protistology, 48, 103–123.
Bruen, T. C., Philippe, H., & Bryant, D. (2006). A simple and robust statistical test for detecting the presence of recombination. Genetics, 172, 2665–2681.
Burke, J. M., Belfort, M., Cech, T. R., Davies, R. W., Schweyen, R. J., Shub, D. A., Szostak, J. W., & Tabak, H. F. (1987). Structural conventions for group I introns. Nucleic Acids Research, 15, 7217–7221.
Cannone, J. J., Subramanian, S., Schnare, M. N., Collett, J. R., D'Souza, L. M., Du, Y., et al. (2002). The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics, 3, 2.
Clark, J. (1993). Didymium iridis reproductive systems: additions and meiotic drive. Mycologia, 85, 764–768.
Clark, J. (1995). Myxomycete reproductive systems: additional information. Mycologia, 87, 779–786.
Clark, J. (2000). The species problem in the myxomycetes. Stapfia, 73, 39–53.
Clark, J., & Haskins, E. F. (2010). Reproductive systems in the myxomycetes: a review. Mycosphere, 1, 337–353.
Clark, J., & Haskins, E. F. (2011). Principles and protocols for genetical study of myxomycete reproductive systems and plasmodial coalescence. Mycosphere, 2, 487–496.
Clark, J., & Haskins, E. F. (2012). Plasmodial incompatibility in the myxomycetes: a review. Mycosphere, 3, 131–141.
Clark, J., & Haskins, E. F. (2013). The nuclear reproductive cycle in the myxomycetes: a review. Mycosphere, 4, 233–248.
Clark, J., Schnittler, M., & Stephenson, S. L. (2002). Biosystematics of the myxomycete Arcyria cinerea. Mycotaxon, 82, 343–346.
Collins, O. N. R. (1975). Mating types in five isolates of Physarum polycephalum. Mycologia, 67, 98–107.
Collins, O. N. R. (1979). Myxomycete biosystematics: some recent developments and future research opportunities. Botanical Review, 45, 145–201.
Collins, O. N. R. (1981). Myxomycete genetics, 1960–1981. Journal of the Elisha Mitchell Scientific Society, 97, 101–125.
Dalgleish, D. (2014). Contexture. Excel tips, tutorials, and videos. Excel latitude and longitude calculations. http://www.contextures.com/excellatitudelongitude.html. Accessed 2 Jan 2014.
DePriest, P. T. (1993). Small subunit rDNA variation in a population of lichen fungi due to optional group-I introns. Gene, 134, 67–74.
Dujon, B. (1989). Group I introns as mobile genetic elements: facts and mechanistic speculations—a review. Gene, 82, 91–114.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797.
Edgell, D. R., Chalamcharla, V. R., & Belfort, M. (2011). Learning to live together: mutualism between self-splicing introns and their hosts. BMC Biology, 9, 22.
Elde, M., Haugen, P., Willassen, N. P., & Johansen, S. (1999). I-NjaI, a nuclear intron-encoded homing endonuclease from Naegleria, generates a pentanucleotide 3' cleavage-overhang within a 19 base-pair partially symmetric DNA recognition site. European Journal of Biochemistry, 259, 281–288.
Fenchel, T., & Finlay, B. J. (2004). The ubiquity of small species: patterns of local and global diversity. BioScience, 54, 777–784.
Ferris, P. J., Vogt, V. M., & Truitt, C. L. (1983). Inheritance of extrachromosomal rDNA in Physarum polycephalum. Molecular and Cellular Biology, 3, 635–642.
Fiore-Donno, A. M., Berney, C., Pawlowski, J., & Baldauf, S. L. (2005). Higher-order phylogeny of plasmodial slime molds (Myxogastria) based on elongation factor 1-A and small subunit rRNA gene sequences. Journal of Eukaryotic Microbiology, 52, 201–210.
Fiore-Donno, A. M., Meyer, M., Baldauf, S. L., & Pawlowski, J. (2008). Evolution of dark-spored Myxomycetes (slime-molds): molecules versus morphology. Molecular Phylogenetics and Evolution, 46, 878–889.
Fiore-Donno, A. M., Nikolaev, S. I., Nelson, M., Pawlowski, J., Cavalier-Smith, T., & Baldauf, S. L. (2010). Deep phylogeny and evolution of slime moulds (Mycetozoa). Protist, 161, 55–70.
Fiore-Donno, A. M., Novozhilov, Y. K., Meyer, M., & Schnittler, M. (2011). Genetic structure of two protist species (Myxogastria, Amoebozoa) suggests asexual reproduction in sexual amoebae. PLoS ONE, 6, e22872.
Fiore-Donno, A. M., Kamono, A., Meyer, M., Schnittler, M., Fukui, M., & Cavalier-Smith, T. (2012). 18S rDNA phylogeny of Lamproderma and allied genera (Stemonitales, Myxomycetes, Amoebozoa). PLoS ONE, 7, e35359.
Fiore-Donno, A. M., Clissmann, F., Meyer, M., Schnittler, M., & Cavalier-Smith, T. (2013). Two-gene phylogeny of bright-spored Myxomycetes (slime moulds, superorder Lucisporidia). PLoS ONE, 8, e62586.
Flick, K. E., Jurica, M. S., Monnat, R. J., Jr., & Stoddard, B. L. (1998). DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature, 394, 96–101.
Flowers, J. M., Li, S. I., Stathos, A., Saxer, G., Ostrowski, E. A., Queller, D. C., et al. (2010). Variation, sex, and social cooperation: molecular population genetics of the social amoeba Dictyostelium discoideum. PLoS Genetics, 6, e1001013.
Frommlet, J. C., & Iglesias-Rodríguez, M. D. (2008). Microsatellite genotyping of single cells of the dinoflagellate species Lingulodinium polyedrum (Dinophyceae): a novel approach for marine microbial population genetic studies. Journal of Phycology, 44, 1116–1125.
Goddard, M. R., & Burt, A. (1999). Recurrent invasion and extinction of a selfish gene. Proceedings of the National Academy of Sciences of the United States of America, 96, 13880–13885.
Goddard, M. R., Greig, D., & Burt, A. (2001). Outcrossed sex allows a selfish gene to invade yeast populations. Proceedings of the Royal Society of London B, 268, 2537–2542.
Goddard, M. R., Leigh, J., Roger, A. J., & Pemberton, A. J. (2006). Invasion and persistence of a selfish gene in the Cnidaria. PLoS ONE, 1, e3.
Gogarten, J. P., & Hilario, E. (2006). Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements. BMC Evolutionary Biology, 6, 94.
Goodman, E. M. (1980). Physarum polycephalum: a review of a model system using a structure-function approach. International Review of Cytology, 63, 1–58.
Gott, J. M., Visomirski, L. M., & Hunter, J. L. (1993). Substitutional and insertional RNA editing of the cytochrome c oxidase subunit 1 mRNA of Physarum polycephalum. The Journal of Biological Chemistry, 268, 25483–25486.
Gray, W. D., & Alexopoulos, C. J. (1968). Biology of the Myxomycetes. New York: The Ronald Press Co.
Guindon, S., & Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology, 52, 696–704.
Hafez, M., & Hausner, G. (2012). Homing endonucleases: DNA scissors on a mission. Genome, 55, 553–569.
Haskins, E. F., & Wrigley de Basanta, D. (2008). Methods of agar culture of Myxomycetes: an overview. Revista Mexicana de Micologia, 27, 1–7.
Haubold, B., & Hudson, R. R. (2000). LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics, 16, 847–848.
Haugen, P., Huss, V. A. R., Nielsen, H., & Johansen, S. (1999). Complex group-I introns in nuclear SSU rDNA of red and green algae: evidence of homing-endonuclease pseudogenes in the Bangiophyceae. Current Genetics, 36, 345–353.
Haugen, P., De Jonckheere, J. F., & Johansen, S. (2002). Characterization of the self-splicing products of two complex Naegleria LSU rDNA group I introns containing homing endonuclease genes. European Journal of Biochemistry, 269, 1641–1649.
Haugen, P., Coucheron, D. H., Rønning, S. B., Haugli, K., & Johansen, S. (2003). The molecular evolution and structural organization of self-splicing group I introns at position 516 in nuclear SSU rDNA of myxomycetes. Journal of Eukaryotic Microbiology, 50, 283–292.
Haugen, P., Reeb, V., Lutzoni, F., & Bhattacharya, D. (2004). The evolution of homing endonuclease genes and group I introns in nuclear rDNA. Molecular Biology and Evolution, 21, 129–140.
Haugen, P., Simon, D. M., & Bhattacharya, D. (2005a). The natural history of group I introns. Trends in Genetics, 21, 111–119.
Haugen, P., Wikmark, O. G., Vader, A., Coucheron, D. H., Sjøttem, E., & Johansen, S. D. (2005b). The recent transfer of a homing endonuclease gene. Nucleic Acids Research, 33, 2734–2741.
Hausner, G., Hafez, M., & Edgell, D. R. (2014). Bacterial group I introns: mobile RNA catalysts. Mobile DNA, 5, 8.
Hedberg, A., & Johansen, S. D. (2013). Nuclear group I introns in self-splicing and beyond. Mobile DNA, 4, 17.
Hibbett, D. S. (1996). Phylogenetic evidence for horizontal transmission of group I introns in the nuclear ribosomal DNA of mushroom-forming fungi. Molecular Biology and Evolution, 13, 903–917.
Hood, G. M. (2010). PopTools version 3.2.5. http://www.poptools.org. Accessed 1 April 2014.
Horton, T. L., & Landweber, L. F. (2000). Evolution of four types of RNA editing in myxomycetes. RNA, 6, 1339–1346.
Huson, D. H., & Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution, 23, 254–267.
Johansen, S., Johansen, T., & Haugli, F. (1992). Structure and evolution of myxomycete nuclear group I introns: a model for horizontal transfer by intron homing. Current Genetics, 22, 297–304.
Johansen, S., Embley, T. M., & Willassen, N. P. (1993). A family of nuclear homing endonucleases. Nucleic Acids Research, 21, 4405.
Johansen, S., Elde, M., Vader, A., Haugen, P., Haugli, K., & Haugli, F. (1997). In vivo mobility of a group I twintron in nuclear ribosomal DNA of the myxomycete Didymium iridis. Molecular Microbiology, 24, 737–745.
Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780.
Koufopanou, V., Goddard, M. R., & Burt, A. (2002). Adaptation for horizontal transfer in a homing endonuclease. Molecular Biology and Evolution, 19, 239–246.
Lado, C. (2014). An on line nomenclatural information system of Eumycetozoa. 2005–2014. http://www.nomen.eumycetozoa.com. Accessed 8 April 2014.
Lahr, D. J. G., Grant, J., Nguyen, T., Lin, J. H., & Katz, L. A. (2011a). Comprehensive phylogenetic reconstruction of Amoebozoa based on concatenated analyses of SSU rDNA and actin genes. PLoS ONE, 6, e22780.
Lahr, D. J. G., Parfrey, L. W., Mitchell, E. A., Katz, L. A., & Lara, E. (2011b). The chastity of amoebae: re-evaluating evidence for sex in amoeboid organisms. Proceedings of the Royal Society of London B, 278, 2081–2090.
Lambowitz, A. M., & Belfort, M. (1993). Introns as mobile genetic elements. Annual Review of Biochemistry, 62, 587–622.
Lazo, W. R. (1961). Growth of green algae with myxomycete plasmodia. The American Midland Naturalist Journal, 65, 381–383.
Li, Z., & Zhang, Y. (2005). Predicting the secondary structures and tertiary interactions of 211 group I introns in IE subgroup. Nucleic Acids Research, 33, 2118–2128.
Lieb, B. (2014). PCR Additives. http://www.staff.uni-mainz.de/lieb/additiva.html. Accessed 6 March 2014.
Linne, C. (1792). Systema Naturae. Tom. II. Pars II. listed under the name Stemonitis. 1467–1470.
Lundblad, E. W., Einvik, C., Rønning, S., Haugli, K., & Johansen, S. (2004). Twelve group I introns in the same pre-rRNA transcript of the myxomycete Fuligo septica: RNA processing and evolution. Molecular Biology and Evolution, 21, 1283–1293.
Machouart, M., Lacroix, C., Bui, H., Feuilhade de Chauvin, M., Derouin, F., & Lorenzo, F. (2004). Polymorphisms and intronic structures in the 18S subunit ribosomal RNA gene of the fungi Scytalidium dimidiatum and Scytalidium hyalinum. Evidence of an IC1 intron with an His-Cys endonuclease gene. FEMS Microbiology Letters, 238, 455–467.
Marchler-Bauer, A., Zheng, C., Chitsaz, F., Derbyshire, M. K., Geer, L. Y., Geer, R. C., et al. (2013). CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Research, 41, D348–D352.
Martin, G. W., & Alexopoulos, C. J. (1969). The Myxomycetes. Iowa City: Iowa Univ. Press.
Michel, F., & Westhof, E. (1990). Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. Journal of Molecular Biology, 216, 585–610.
Milne, I., Lindner, D., Bayer, M., Husmeier, D., McGuire, G., Marshall, D. F., & Wright, F. (2009). TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics, 25, 126–127.
Milstein, D., Oliveira, M. C., Martins, F. M., & Matioli, S. R. (2008). Group I introns and associated homing endonuclease genes reveals a clinal structure for Porphyra spiralis var. amplifolia (Bangiales, Rhodophyta) along the eastern coast of South America. BMC Evolutionary Biology, 8, 308.
Moriyama, Y., & Kawano, S. (2010). Maternal inheritance of mitochondria: multipolarity, multiallelism and hierarchical transmission of mitochondrial DNA in the true slime mold Physarum polycephalum. Journal of Plant Research, 123, 139–148.
Müller, K. M., Cannone, J. J., Gutell, R. R., & Sheath, R. G. (2001). A structural and phylogenetic analysis of the group IC1 introns in the order Bangiales (Rhodophyta). Molecular Biology and Evolution, 18, 1654–1667.
Muscarella, D. E., & Vogt, V. M. (1989). A mobile group I intron in the nuclear rDNA of Physarum polycephalum. Cell, 56, 443–454.
Muscarella, D. E., & Vogt, V. M. (1993). A mobile group I intron from Physarum polycephalum can insert itself and induce point mutations in the nuclear ribosomal DNA of Saccharomyces cerevisiae. Molecular and Cellular Biology, 13, 1023–1033.
Muscarella, D. E., Ellison, E. L., Ruoff, B. M., & Vogt, V. M. (1990). Characterization of I-Ppo, an intron-encoded endonuclease that mediates homing of a group I intron in the ribosomal DNA of Physarum polycephalum. Molecular and Cellular Biology, 10, 3386–3396.
Nandipati, S. C., Haugli, K., Coucheron, D. H., Haskins, E. F., & Johansen, S. D. (2012). Polyphyletic origin of the genus Physarum (Physarales, Myxomycetes) revealed by nuclear rDNA mini-chromosome analysis and group I intron synapomorphy. BMC Evolutionary Biology, 12, 166.
Nannenga-Bremekamp, N. B. (1991). A guide to temperate Myxomycetes (Feest A, Burgraff E: De Nederlandse Myxomyceten, Engl. transl.). Bristol: Biopress Lim.
Neubert, H., Nowotny, W., & Baumann, K. (1993). Die Myxomyceten Deutschlands und des angrenzenden Alpenraumes unter besonderer Berücksichtigung Österreichs. Band 1. Ceratiomyxales, Echinosteliales, Liceales, Trichiales. Gomaringen: Baumann Verl.
Nielsen, H., & Johansen, S. D. (2009). Group I introns: moving in new directions. RNA Biology, 6, 375–383.
Nikoh, N., & Fukatsu, T. (2001). Evolutionary dynamics of multiple group I introns in nuclear ribosomal RNA genes of endoparasitic fungi of the genus Cordyceps. Molecular Biology and Evolution, 18, 1631–1642.
Novozhilov, Y. K., Okun, M. V., Erastova, D. A., Shchepin, O. N., Zemlyanskaya, I. V., García-Carvajal, E., & Schnittler, M. (2013a). Description, culture and phylogenetic position of a new xerotolerant species of Physarum. Mycologia, 105, 1535–1546.
Novozhilov, Y. K., Schnittler, M., Erastova, D. A., Okun, M. V., Schepin, O. N., & Heinrich, E. (2013b). Diversity of nivicolous myxomycetes of the Teberda State Biosphere Reserve (Northwestern Caucasus, Russia). Fungal Diversity, 59, 109–130.
Pawlowski, J., Audic, S., Adl, S., Bass, D., Belbahri, L., Berney, C., et al. (2012). CBOL protist working group: barcoding eukaryotic richness beyond the Animal, Plant, and Fungal kingdoms. PLoS Biology, 10, e1001419.
Persoon, C. H. (1794). Neuer Versuch einer systematischen Eintheilung der Schwämme. Neues Magazin für die Botanik in ihrem ganzen Umfange, 1, 63–128.
Poulain, M., Meyer, M., & Bozonnet, J. (2011). Les myxomycetes. Delémont: Féd. Mycol. Bot. Dauphiné-Savoie.
Rätzel, V., Ebeling, B., Hoffman, X. K., Tesmer, J., & Marwan, W. (2013). Physarum polycephalum mutants in the photocontrol of sporulation display altered patterns in the correlated expression of developmentally regulated genes. Development Growth and Differentiation, 55, 247–259.
Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542.
Sauer, H. W. (1982). Developmental Biology of Physarum. Cambridge: Cambridge University Press.
Schnittler, M., & Mitchell, D. W. (2000). Species diversity in Myxomycetes based on the morphological species concept—a critical examination. Stapfia, 73, 55–62.
Schnittler, M., & Tesmer, J. (2008). A habitat colonisation model for spore-dispersed organisms—does it work with eumycetozoans? Mycological Research, 112, 697–707.
Schnittler, M., Novozhilov, Y. K., Romeralo, M., Brown, M., & Spiegel, F. W. (2012). Myxomycetes and Myxomycete-like organisms. In F. W. Stuttgart (Ed.), Englers Syllabus of Plant Families. Volume 4. 13th edition (pp. 40–88). Bornträger.
Smirnov, A. V., Chao, E., Nassonova, E. S., & Cavalier-Smith, T. (2011). A revised classification of naked lobose amoebae (Amoebozoa: Lobosa). Protist, 162, 545–570.
STAR. (2014). StarORF. Software Tools for Academics and Researchers. http://star.mit.edu/orf. Accessed 25 March 2014.
Stephenson, S. L., Schnittler, M., & Novozhilov, Y. K. (2008). Myxomycete diversity and distribution from the fossil record to the present. Biodiversity and Conservation, 17, 285–301.
Tamura, K., Nei, M., & Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences of the United States of America, 101, 11030–11035.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739.
Tanabe, Y., Yokota, A., & Sugiyama, J. (2002). Group I introns from Zygomycota: evolutionary implications for the fungal IC1 intron subgroup. Journal of Molecular Biology, 54, 692–702.
Torres-Machorro, A. L., Hernández, R., Cevallos, A. M., & López-Villaseñor, I. (2010). Ribosomal RNA genes in eukaryotic microorganisms: witnesses of phylogeny? FEMS Microbiology Reviews, 34, 59–86.
Traphagen, S. J., Dimarco, M. J., & Silliker, M. E. (2010). RNA editing of 10 Didymium iridis mitochondrial genes and comparison with the homologous genes in Physarum polycephalum. RNA, 16, 828–838.
Urich, T., Lanzén, A., Qi, J., Huson, D. H., Schleper, C., & Schuster, S. C. (2008). Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS ONE, 3, e2527.
Vader, A., Nielsen, H., & Johansen, S. (1999). In vivo expression of the nucleolar group I intron-encoded I-DirI homing endonuclease involves the removal of a spliceosomal intron. The EMBO Journal, 18, 1003–1013.
Walker, L. M., Dewsbury, D. R., Parks, S. S., Winsett, K. E., & Stephenson, S. L. (2011). The potential use of mitochondrial cytochrome c oxidase I for barcoding myxomycetes. In VII International Congress on Systematics and Ecology of Myxomycetes: 11–16 September 2011; Recife, Brazil (pp. 138).
Wikmark, O. G., Haugen, P., Haugli, K., & Johansen, S. D. (2007a). Obligatory group I introns with unusual features at positions 1949 and 2449 in nuclear LSU rDNA of Didymiaceae myxomycetes. Molecular Phylogenetics and Evolution, 43, 596–604.
Wikmark, O. G., Haugen, P., Lundblad, E. W., Haugli, K., & Johansen, S. D. (2007b). The molecular evolution and structural organization of group I introns at position 1389 in nuclear small subunit rDNA of myxomycetes. Journal of Eukaryotic Microbiology, 54, 49–56.
Xu, C., Wang, C., Sun, X., Zhang, R., Gleason, M. L., Eiji, T., & Sun, G. (2013). Multiple group I introns in the small-subunit rDNA of Botryosphaeria dothidea: implication for intraspecific genetic diversity. PLoS ONE, 8, e67808.
Yokoyama, E., Yamagishi, K., & Hara, A. (2002). Group-I intron containing a putative homing endonuclease gene in the small subunit ribosomal DNA of Beauveria bassiana IFO 31676. Molecular Biology and Evolution, 19, 2022–2025.
Zhou, Y., Lu, C., Wu, Q. J., Wang, Y., Sun, Z. T., Deng, J. C., & Zhang, Y. (2008). GISSD: group I intron sequence and structure database. Nucleic Acids Research, 36, D31–D37.
Acknowledgments
This research was supported by the Deutsche Forschungsgemeinschaft (DFG) to MS (SCHN 1080/2-1). The authors owe thanks for technical support to Anja Klahr, Greifswald. Fieldwork in the Bavarian Forest National Park was supported by Claus Bässler from the research unit of the park administration. We wish to thank Anna Maria Fiore-Donno for suggesting four primer sequences. For loans of specimens of T. varia, we are indebted to A.M. Fiore-Donno, Germany; Myriam de Haan, Belgium; Hans van Hooff, Netherlands; Yuri K. Novozhilov, Russia; Anna Ronikier, Poland; and Jos Van Roy, Belgium.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure S1
Position of Trichia varia in a Bayesian majority-rule consensus tree of the complete SSU gene for bright-spored myxomycetes with Ceratiomyxa fruticulosa as outgroup. Support values are shown for nodes with both Bayesian posterior probability >0.70 and bootstrap replicates >50. (PDF 343 kb)
Supplementary Figure S2
ML tree constructed from exon parts of complete SSU for 66 accessions of Trichia varia. Support values are shown for nodes with both Bayesian posterior probability >0.70 and bootstrap replicates >50. (PDF 269 kb)
Supplementary Table S1
Specimens used in this study and their assignment to genotypes with GenBank accession numbers. (DOC 563 kb)
Supplementary Table S2
Known SSU sequences of myxomycetes with group I intron S956. (DOC 92 kb)
Supplementary Database S1 (Microsoft Excel 2013)
Locality descriptions for all specimens investigated in this study. Geographic coordinates are given in the format dd°mm'ss.s", elevations in meter above sea level. (XLS 109 kb)
Rights and permissions
About this article
Cite this article
Feng, Y., Schnittler, M. Sex or no sex? Group I introns and independent marker genes reveal the existence of three sexual but reproductively isolated biospecies in Trichia varia (Myxomycetes). Org Divers Evol 15, 631–650 (2015). https://doi.org/10.1007/s13127-015-0230-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-015-0230-x