Abstract
Hawaii is one of the most isolated archipelagos in the world, situated about 4,000 km from the nearest continent, and never connected with continental land masses. Two Hawaiian endemic blueberries, Vaccinium calycinum and V. reticulatum, are infected by Exobasidium species previously recognized as Exobasidium vaccinii. However, because of the high host-specificity of Exobasidium, it seems unlikely that the species infecting Vaccinium calycinum and V. reticulatum belongs to Exobasidium vaccinii, which in the current circumscription is restricted to Vaccinium vitis-idaea. We collected a fresh specimen of Exobasidium on Vaccinium reticulatum in Haleakala National Park, and analysed it by a combined morphological and molecular approach. The morphological and phylogenetic analyses (based on the LSU and the concatenated ITS+LSU sequence data) showed that Exobasidium on Vaccinium reticulatum belongs to an undescribed species, distinct from Exobasidium vaccinii and any other species. The name Exobasidium darwinii is proposed for this novel taxon. This species is characterized among others by the production of peculiar witches’ brooms with bright red leaves on the infected branches of Vaccinium reticulatum. Relevant characters of Exobasidium darwinii are described and illustrated, additionally phylogenetic relationships of the new species are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Exobasidium Woronin is the largest genus within the Exobasidiales with 178 epithets included in the Index Fungorum (http://www.indexfungorum.org/Names/Names.asp). Some of these names are invalidly published, others are synonymous or represent species of other genera (e.g., Arcticomyces Savile, Exobasidiellum Donk, Kordyana Racib., Laurobasidium Jülich, Muribasidiospora Kamat & Rajendren). Yet the genus is poorly known and, except for Europe, there is no comprehensive study of Exobasidium. In the past, Exobasidium species were described based on supposed host specificity, but this concept was later abandoned by Burt (1915) and Savile (1959), who lumped together many species, especially within Exobasidium vaccinii (Fuckel) Woronin and Exobasidium vaccinii-uliginosi Boud. The high level of host specialization was again postulated for European Exobasidium species by Nannfeldt (1981) on the basis of a detailed analysis of morphological characters correlated with host plant and physiological data (the latter elaborated by Sundström 1964). His ideas were basically supported by molecular analyses of Blanz and Oberwinkler (1983), Blanz and Döring (1995) and Begerow et al. (2002), although in this latter study support for most branches was low and obtained resolution was not fully satisfactorily for conclusions about evolutionary relationships within both Exobasidium and the Exobasidiales.
Located in the middle of the Pacific Ocean, Hawaii is one of the most isolated archipelagos in the world. The islands are situated about 4,000 km from the nearest continent and have never been connected with continental land masses. The indigenous flowering plants of Hawaii include 1,029 species, of which ca. 89% are endemic to the islands (Sakai et al. 2002) likely harbouring many undescribed pathogenic fungi that are usually strictly correlated with host plants. Two Hawaiian endemic blueberries, Vaccinium calycinum Sm. and V. reticulatum Sm. (incl. V. peleanum Skottsb.), were reported as hosts for Exobasidium species producing peculiar witches’ brooms with bright red leaves on infected branches (Gardner 1985). Although the causal agent was first considered as a mycoplasma-like organism, it soon became evident that it is rather a species of Exobasidium, which has been referred to as Exobasidium vaccinii but with uncertainty whether it is native or alien on Hawaiian blueberries (Gardner 1985). However, in the light of the high specificity of Exobasidium, usually to the level of host species, rarely to a group of closely allied species (Nannfeldt 1981), it seems unlikely that the species infecting Vaccinium calycinum and V. reticulatum belongs to Exobasidium vaccinii. That latter species is quite a different fungus, producing localized, conspicuous and much thickened concavities on leaves of Vaccinium vitis-idaea L. Only occasionally it may cause deformations of all shoot-tips (Nannfeldt 1981), which however are never as strong as on the Hawaiian blueberries. Therefore, we hypothesize that Exobasidium on Vaccinium calycinum and V. reticulatum represents in fact one (or even two) distinct species. To test this hypothesis, we collected a fresh specimen of Exobasidium on Vaccinium reticulatum in Haleakala National Park, and analysed it by a combined morphological and molecular approach. Unfortunately, the corresponding specimen of Exobasidium on Vaccinium calycinum has not hitherto been re-collected.
Materials and methods
The Exobasidium specimen infecting Vaccinium reticulatum was collected in Haleakala National Park in Hawaii on 17 December 2007. This specimen was used in subsequent morphological and phylogenetic analyses. Information on the specimens of Exobasidium arescens Nannf. and E. rostrupii Nannf. sequenced for the present study is given in Table 1.
Morphological analyses
The macroscopic symptoms of host infection and microscopic features of Exobasidium on Vaccinium reticulatum were studied using dried herbarium material. The terminology used in the species description and in the text follows Nannfeldt (1981). Morphological observations and measurements of basidia, basidiospores and conidia were made by standard light and phase contrast microscopy (LM) of the material mounted in 3% KOH. A Nikon Eclipse 80i light microscope was used for LM studies. Thirty basidiospores and conidia were measured from the studied collection, using NIS-Elements BR 3.0 imaging software. LM micrographs were taken with a Nikon DS-Fi1 camera. The observations of basidia were supplemented by scanning electron microscopy (SEM). For this purpose, a fragment of infected leaf was glued to the carbon tab and fixed to an aluminium stub with double-sided transparent tape. The sample was sputter-coated with carbon using a Cressington sputter-coater and viewed with a Hitachi S-4700 scanning electron microscope, at 10 kV and 9,000 nA, with a working distance of ca. 12 mm. SEM micrographs were taken in the Laboratory of Field Emission Scanning Electron Microscopy and Microanalysis at the Institute of Geological Sciences of the Jagiellonian University, Kraków (Poland).
Molecular analyses
Genomic DNA was isolated from the herbarium specimens. For methods of isolation and crushing of fungal material, DNA extraction, amplification, purification of PCR products, sequencing, and processing of the raw data, see Lutz et al. (2004). We determined base sequences of the ITS1/2 region of the nuc-rDNA including the 5.8 S rDNA (ITS) and of the 5´-end of the nuc-LSU rDNA including the domains D1/D2 (LSU). The ITS was amplified using the primer pair ITS1f and ITS4 (Gardes and Bruns 1993). The LSU was amplified using the primer pair NL1 and NL4 (O’Donnell 1992, 1993). For amplification of both regions, we adjusted the annealing temperature to 45°C. DNA sequences prepared in the course of this study were deposited in GenBank; accession numbers are given in Table 1, and Figs. 1 and 2.
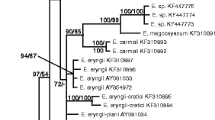
Bayesian inference of phylogenetic relationships within the sampled Exobasidiales: Markov chain Monte Carlo analysis of an alignment of LSU base sequences using the GTR + I + G model of DNA substitution with gamma distributed substitution rates and estimation of invariant sites, random starting trees and default starting parameters of the DNA substitution model. A 70% majority-rule consensus tree computed from 18,000 trees that were sampled after the process had reached stationarity is shown. The topology was rooted with Tilletiopsis pallescens (AB178241, AB178262, AB178263). Numbers on branches before slashes are estimates for a posteriori probabilities, numbers on branches after slashes are percentage bootstrap values of 1,000 replicates. Branch lengths were averaged over the sampled trees. They are scaled in terms of expected numbers of nucleotide substitutions per site. E. = Exobasidium, T. = Tilletiopsis
Bayesian inference of phylogenetic relationships within the sampled Exobasidium species: Markov chain Monte Carlo analysis of a concatenated ITS+LSU alignment using the GTR+I+G model of DNA substitution with gamma distributed substitution rates and estimation of invariant sites, random starting trees and default starting parameters of the DNA substitution model. A 70% majority-rule consensus tree computed from 18,000 trees that were sampled after the process had reached stationarity is shown. The topology was rooted with Exobasidium kishianum (AB180353/AB177577) and E. vaccinii (AB180362/AB177560). Numbers on branches before slashes are estimates for a posteriori probabilities, numbers on branches after slashes are percentage bootstrap values of 1,000 replicates. Branch lengths were averaged over the sampled trees. They are scaled in terms of expected numbers of nucleotide substitutions per site. E. = Exobasidium, Eu. = Eubotryoides
To obtain a hypothesis on the phylogenetic position of the specimen infecting Vaccinium reticulatum, we followed two strategies: (1) we analysed its LSU sequence together with sequences of most Exobasidium species available in GenBank, including sequences of all available species growing on Vaccinium L. and all available species producing witches’ brooms, respectively, and sequences of selected representatives of other genera of Exobasidiales (GenBank accession numbers are given in Fig. 1), and (2) we analysed the concatenated ITS+LSU sequences of the specimen from Hawaii within a dataset reduced to the closest relatives according to the results of the LSU analyses (GenBank accession numbers are given in Fig. 2). The species and sequences used in phylogenetic analyses are listed in Table 1.
Sequences were aligned for both datasets with MAFFT 6.611 (Katoh et al. 2002, 2005; Katoh and Toh 2008) using the L-INS-i option. Both alignments [length: 559 bp (LSU), 1,106 bp (ITS+LSU); variable sites: 185 (LSU), 163 (ITS+LSU)] were used throughout their length. We avoided both manipulation of the alignment by hand and manual exclusion of any positions as recommended by Giribet and Wheeler (1999) and Gatesy et al. (1993), respectively.
For phylogenetic analyses, we used both neighbour-joining analysis and a Bayesian approach. For neighbour-joining analysis, the data were first analysed with Modeltest 3.7 (Posada and Crandall 1998) to find the most appropriate model of DNA substitution. For the LSU dataset, the hierarchical likelihood ratio test proposed the SYM+I+G DNA substitution model, while for the ITS+LSU dataset, the GTR+I+G DNA substitution model was proposed. Bootstrap values were calculated for 1,000 replicates. For Bayesian analysis, we used for both datasets a Bayesian approach of phylogenetic inference using a Markov chain Monte Carlo (MCMC) technique as implemented in the computer program MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). In each run, four incrementally heated simultaneous Markov chains were run over 2,000,000 generations using the general time reversible model of DNA substitution with gamma distributed substitution rates and estimation of invariant sites, random starting trees and default starting parameters of the DNA substitution model as recommended by Huelsenbeck and Rannala (2004). Trees were sampled every 100th generation resulting in an overall sampling of 20,001 trees. From these, the first 2,001 trees were discarded (burnin = 2,001). The trees sampled after the process had reached stationarity (18,000 trees) were used to compute a 70% majority rule consensus tree to obtain estimates for the a posteriori probabilities of groups of species. This Bayesian approach of phylogenetic analysis was repeated four times to test the independence of the results from topological priors (Huelsenbeck et al. 2002). Trees resulting from the LSU analyses were rooted with Tilletiopsis pallescens Gokhale (AB178241, AB178262, AB178263), trees resulting from the ITS+LSU analyses were rooted with Exobasidium kishianum Nagao & Ezuka (AB180353/AB177577) and E. vaccinii (AB180362/AB177560).
Clarification of species name in GenBank sequences attributed to Exobasidium sawadae
GenBank includes five sequences (AB180353, AB178241, AB177577, AB177555, AB180354) stored under the name “Exobasidium sawadae Nagao et Ezuka”. This is an apparently provisional name, which was never published by the authors because it would cause the creation of later homonym. The name Exobasidium sawadae G. Yamada 1919 is already available for a completely different species. Recently, the fungus included in GenBank as “Exobasidium sawadae Nagao et Ezuka” was described by Nagao et al. (2006) under the name Exobasidium kishianum. Thus, we used this latter name for these sequences, however, with one exception: our molecular analyses clearly showed that “Exobasidium sawadae” AB178241 in fact represents Tilletiopsis pallescens (see results and discussion).
Results
Molecular phylogenetic analyses
For both the LSU and the concatenated ITS+LSU dataset, all runs of Bayesian phylogenetic analyses that were performed yielded consistent topologies which were congruent to the results of the neighbour-joining analysis in respect to well-supported branchings (a posteriori probability greater than 60%, bootstrap support greater than 73%). For both datasets, we present the consensus tree of one run of Bayesian phylogenetic analyses to illustrate the results (Figs. 1, 2). Estimates for a posteriori probabilities are indicated on branches before slashes, bootstrap values from the neighbour-joining analysis are indicated after slashes.
The analyses of the LSU dataset placed the Exobasidium specimen infecting Vaccinium reticulatum from Hawaii as a sister taxon of E. inconspicuum Nagao & Ezuka within a well-supported group of four more Exobasidium species (E. arescens, E. bisporum Sawada ex Ezuka, E. pachysporum Nannf., E. rostrupii). Within that group, only the monophyly of E. rostrupii and E. arescens received considerable support values. The relationship between that group of species and most of the other Exobasidium species sampled remained unresolved using LSU sequences. Moreover, the relation between the sampled Exobasidium species and Muribasidiospora indica Kamat & Rajendren, Arcticomyces warmingii (Rostr.) Savile (both belonging to the Exobasidiaceae), Kordyana tradescantiae (Pat.) Racib. (Brachybasidiaceae), and Graphiola phoenicis (Moug. ex Fr.) Poit. (Graphiolaceae) was not resolved. However, the cryptobasidiaceous species Clinoconidium bullatum Syd. and Laurobasidium lauri (Geyl.) Jülich were revealed as sister taxon to all other Exobasidiales sampled. The sequence “Exobasidium kishianum AB178241” clustered together with the Tilletiopsis pallescens sequences AB178262 and AB178263 (Fig. 1). A blast search (Altschul et al. 1997) resulted in 100% identity of the sequence “Exobasidium kishianum AB178241” with the sequences of Tilletiopsis pallescens AB178263, AB178262, AJ235329, AJ235293, AJ235292, AJ235291, and of T. albescens Gokhale AJ235290.
In contrast to the analyses of the LSU dataset, those of the ITS+LSU dataset resolved the relationships within the cluster of six species mentioned above with good to moderate support. The Exobasidium specimen infecting Vaccinium reticulatum from Hawaii clustered as moderately supported sister taxon of E. bisporum in a well supported group together with E. pachysporum (Fig. 2).
Taxonomy
Exobasidium darwinii M. Piątek & M. Lutz, sp. nov. (Figs. 3 and 4)
Exobasidium darwinii sp. nov. on Vaccinium reticulatum (all from holotype: KRAM F-47124): a infected plants in natural habitat, b infected branches of the host plant with uniformly bright red, not hypertrophied and not enlarged leaves, c witches’ broom, d basidium emerging through stomatum, as seen by SEM, e basidium with four sterigmata, as seen by SEM, f–h basidia, as seen by LM, i–o variability of basidiospores, note the septa visible on j and m, as seen by LM, p–s bacilliform conidia, as seen by LM. Bars 10 μm (d, f–s), 5 μm (e)
MycoBank no. MB 518226
Etymology
Named after Charles Darwin (1809–1882), whose observations, among others on the oceanic Galapagos Islands, resulted in formulating the evolutionary theory.
Parasitus in Vaccinio reticulato. Infectio systemica. Fungus in planta hospitali per proliferationem continuam parvorum novorum ramulorum conspicuas scopas strigae facit; rami infecti saepe modice deformati, contorti cum intumescentiis associatis. Folia infecta aequaliter pallide rubra ex utraque parte, non hypertrophica et non dilatata, prae foliis validis, hymenium hypophyllum satis tenue, ex basidiis, basidiosporis et conidiis compositum. Basidia singulatim vel in fasciculis per stomata vel directe ex epidermi deleta emergentia, hyalina, cylindrica, 20–30 μm longa, 3.9–5.6 μm lata, apice rotundata et 3–4 sterigmatibus terminata. Sterigmata 2.2–5.5 μm longa et 1.4–1.9 μm in basi lata, ad apicem attenuata. Basidiosporae hyalinae, cylindricae, subcylindricae, musaiformes, saepe in uno apice curvatae, plerumque versus hilum attenuatae 11.5–18.5 × 2.5–3.8 μm, 0–3-septatae, plerumque cum paucis guttulis oleosis, paries glaber. Conidia hyalina, bacilliformia, 5–10 × 1.5–2.5 μm, non septata, paries glaber. Sequentiae acidi nucleici ITS/LSU typi in collectione sequentiarum acidi nucleici NCBI (GenBank) ut FJ896133/FJ866134 depositae sunt.
Type
On Vaccinium reticulatum Sm., U.S.A., Hawaii, Maui, north facing slopes of Haleakala, near Hosmers Grove, elev. ca. 6,800 ft. (= ca. 2,072 m a.s.l.), 17 Dec. 2007, leg. P. Welton (holotype: KRAM F-47124, isotype: TUB 019166).
Parasitic on Vaccinium reticulatum. Infection systemic. Fungus produces conspicuous witches’ brooms on the host plant through continuing proliferation of small young branches; infected twigs often somewhat deformed, contorted and accompanied with swellings. Infected leaves uniformly bright red on both sides, not hypertrophied and not enlarged in comparison with healthy leaves; hymenium hypophyllous, quite delicate, composed of basidia, basidiospores and conidia. Basidia emerging singly or in fascicles through stomata or directly from disintegrated epidermis, hyaline, cylindrical, 20–30 μm long, 3.9–5.6 μm wide, apically rounded and ended with 3–4 sterigmata. Sterigmata 2.2–5.5 μm long and 1.4–1.9 μm wide at the base, tapering toward the tip. Basidiospores hyaline, cylindrical, subcylindrical, musiform, often curved at one end, usually tapering toward the hilum, 11.5–18.5 × 2.5–3.8 μm, 0–3-septate, usually with several oil drops, wall smooth. Conidia hyaline, bacilliform, 5–10 × 1.5–2.5 μm, non-septate, wall smooth. The ITS/LSU hologenetype sequences are deposited in GenBank as FJ896133/FJ896134, respectively.
Distribution Vaccinium reticulatum is an endemic species of the Hawaiian Archipelago occurring most frequently on Maui and Hawai‘i, and rarely on Kaua‘i, O‘ahu and Moloka‘i at elevations from ca. 600–3,650 m a.s.l. (Wagner et al. 1990). The type specimen of Exobasidium darwinii was collected near Hosmers Grove in Haleakala National Park on the Island of Maui. Gardner (1985) on the basis of aerial observations concluded that Exobasidium on Vaccinium reticulatum is common throughout extensive remote areas of the Park. However, it is not excluded that his observations may at least partly refer to the reddish coloration of juvenile leaves. It is desirable that future studies look for the exact distribution of Exobasidium darwinii on the Hawaiian Islands in order to establish an effective conservation management of this unique species.
Discussion
The morphological observations and phylogenetic analyses in the present study show that Exobasidium on Vaccinium reticulatum in the Hawaiian Archipelago belongs to an undescribed species, distinct from Exobasidium vaccinii and any other species. Thus, the name Exobasidium darwinii is proposed for this novel taxon.
Selection of Exobasidium sequences for molecular analyses
GenBank offers quite a lot of sequences of Exobasidium species generated by various authors, including a considerable number of sequences submitted by H. Nagao in 2004. Many Exobasidium sequences in GenBank were apparently taken from cultures linked with permanently preserved corresponding voucher specimens, others were taken directly from voucher specimens and still others from cultures not linked with herbarium specimens. When constructing the phylogenetic trees, we (1) primarily used sequences taken from Exobasidium species on their type hosts (with one exception—in the case of E. bisporum, the sequence from the type host was not available), (2) obtained from specimens in planta (preferably) or cultures linked with corresponding voucher specimens, and (3) avoided (with some exceptions, when other sequences were not available) using sequences taken from cultures not linked with any voucher specimen. The reason for the two latter choices lies in the fact that the macroscopic appearance of symptoms on the host plant is essential for delimiting the species in Exobasidium, while absence of voucher specimens prevents future verification of species identification. It is especially important because the species concept in Exobasidium was and still is not fully understood in many studies. We encourage to employ a similar approach in future molecular studies on the genus Exobasidium. This is in line with similar observation of Lutz et al. (2011) on urocystidalean species.
Not unimportantly, cultures taken from Exobasidium specimens can be contaminated by co-occurring Tilletiopsis Derx species, and, as Begerow et al. (2000) pointed out, it is difficult to obtain a pure culture of Exobasidium. In fact, Boekhout et al. (1995) found that the culture “Exobasidium vexans CBS 24752” (= sequence AJ235288) represents a Tilletiopsis species. Our analyses indicate that the sequence AB178241 of “Exobasidium kishianum MAFF 306199” was also taken from contaminated material and represents Tilletiopsis pallescens. Interestingly, according to GenBank data, the culture MAFF 306199 was obtained from voucher NIAES 10519, which is the holotype of Exobasidium kishianum (Nagao et al. 2006), included in GenBank under the provisional name “Exobasidium sawadae Nagao et Ezuka” (which is not E. sawadae G. Yamada 1919). Thus, the sequence “Exobasidium kishianum AB178241” should not be used in future phylogenetic studies on the genus Exobasidium, or considered as type sequence of Exobasidium kishianum.
The symptoms and infection caused by Exobasidium darwinii
On host plants, Exobasidium species can cause three different kinds of symptoms, depending on the manner of infection: circumscribed, surculicolous and systemic (Nannfeldt 1981). The infection may be monocarpic (annual, ephemeral) when the pathogen completely dies after sporulation, or polycarpic (perennial) when the pathogen survives in infected plants in the form of intercellular mycelium (Nannfeldt 1981). The circumscribed infection is a monocarpic, strictly localized infection forming leaf spots, leaf concavity galls or gall apples entirely surrounded by healthy tissues of the host plant. The surculicolous infection is also monocarpic, but in that case whole annual shoots are penetrated by the pathogen as in the case of the true systemic infection. In contrast to these two monocarpic symptom types, the systemic infection is polycarpic, and whole plants, single shoots or shoot complexes are invaded, depending on the Exobasidium species (Nannfeldt 1981). The main difference between systemic and surculicolous infections lies in the fact that in the systemic infection the mycelium of Exobasidium species survives in the host plant tissues during the winter season, while in the surculicolous infection it does not. This distinction, however, is often difficult to delineate, and even Nannfeldt (1981), who introduced these terms, pointed out that there may be transitional forms in some Exobasidium species. In the light of these data, the symptoms caused by Exobasidium darwinii could be assigned to the polycarpic, systemic infection because the pathogen surely penetrates whole shoot complexes of Vaccinium reticulatum leading to formation of witches’ brooms within which mycelium apparently survives through the years. The infection is, however, probably limited to certain areas of the host plant since witches’ brooms are formed only locally on certain branches while other branches are healthy.
Comparison of Exobasidium darwinii with other Exobasidium species on Vaccinium and other Exobasidium species causing witches’ brooms
Besides Rhododendron L., the members of the genus Vaccinium harbour the largest number of Exobasidium species. Together with Exobasidium darwinii, there are 22 species and one variety described on different Vaccinium species (Gómez and Kisimova-Horovitz 1998a, b; Hotson 1927; Nagao et al. 2006; Nannfeldt 1981; Nickerson 1984; Piepenbring et al. 2010; Raciborski 1909; Ramakrishnan and Ramakrishnan 1949; Savile 1959). Of them, 8 species and 1 variety are clearly circumscribed taxa, while the remaining 14 species are surculicolous or systemic. Exobasidium darwinii belongs to the latter group, having systemic and polycarpic infections. None of the hitherto described species is morphologically similar to E. darwinii because none of them produces distinct witches’ brooms. Exobasidium parvifolii Hotson on Vaccinium parvifolium Sm. and V. ovalifolium Sm. in North America forms perennial galls on the stems from which numerous cylindrical, clavarioid excrescences emerge. Nannfeldt (1981) interpreted these structures as highly disorganized shoots, which, however, cannot be confused with witches’ brooms of Exobasidium darwinii.
The unusual morphological aspect of Exobasidium darwinii is the production of witches’ brooms on the infected host plant (Fig. 3c). Witches’ brooms are produced by phytopathogenic fungi from different, unrelated systematic groups, but were rarely observed in Exobasidium species. To the best of our knowledge, the witches’ brooms were observed only in four species: Exobasidium uvae-ursi (Maire) Juel on Arctostaphylos uva-ursi (L.) Spreng. in Europe and North America, E. disterigmaticola L.D. Gómez & Kisim.-Hor. on Disterigma humboldtii (Kl.) Nied. in Costa Rica, E. nobeyamense Nagao & Ezuka on Rhododendron wadanum Makino in Japan, and E. pentasporium Shirai on Rhododendron obtusum (Lindl.) Planch. var. kaempferi (Planch.) E.H. Wilson (type host) and R. macrosepalum Maxim. in Japan. Additionally, Nannfeldt (1981) mentioned another, presumably undescribed, Exobasidium species on North American Rhododendron macrophyllum D. Don ex G. Don, but this species is not sufficiently known.
All these species are different from Exobasidium darwinii, not only because of other host plant genera but also by metric characters of basidiospores or conidia. The basidiospores are 15–22 × 5–6 μm in E. uvae-ursi (Nannfeldt 1981), 12–21 × 2–5.5 μm in E. nobeyamense (Nagao et al. 2001) and ca. 14.4 × 4 μm in E. pentasporium (Nannfeldt 1981). Basidiospores were not observed in E. disterigmaticola. However, its conidia (12–15.5 × 2.8–3.2 μm; Gómez and Kisimova-Horovitz 1998b) are much larger compared to those of E. darwinii. In many groups of Exobasidium, basidiospore size shows only a small variance between species. Therefore, these differences distinguish these species well. Additionally, in the phylogenetic analyses, E. nobeyamense and E. pentasporium are placed in the clade comprising species infecting members of Rhododendron, well separated from the clade including E. darwinii. The sequences of E. uvae-ursi and E. disterigmaticola are still not available, and thus their phylogenetic position cannot be resolved. However, the morphological differences do not support their conspecificity with E. darwinii.
Hypothesis on the origin of Exobasidium darwinii
Three Hawaiian Vaccinium species: V. calycinum, V. dentatum Sm., V. reticulatum and the Polynesian V. cereum (L.f.) G. Forst. were included in Vaccinium sect. Macropelma (Klotzsch) Hook.f. (Vander Kloet 1993, 1996). Phylogenetic molecular analyses by Powell and Kron (2002) revealed that the Hawaiian Vaccinium species indeed form a monophyletic group that is nested within members of Vaccinium sect. Myrtillus Dumort. Powell and Kron (2002) suggested a boreal-arctic origin of the species instead of a southern Pacific origin and proposed that Hawaiian blueberries should be transferred to sect. Myrtillus. This view was recently accepted by Vander Kloet and Dickinson (2009).
Of the three Hawaiian Vaccinium species, no Exobasidium has so far been observed on V. dentatum, while the specimen on V. calycinum was not available for analysis. In the present study, Exobasidium darwinii on V. reticulatum is a member of a well-supported clade (LSU analysis, Fig. 1; ITS+LSU analysis, Fig. 2) containing Exobasidium species on different Vaccinium species as well as on Eubotryoides grayana (Maxim.) H. Hara var. glabra (Komatsu) H. Hara and var. oblongifolia (Miq.) H. Hara. The specimens used in the analyses come from Japan, Poland and Slovenia. What is interesting to note is that, except the evidently systemic Exobasidium darwinii, all the remaining taxa from this clade are circumscribed, monocarpic species, all producing merely leaf spots and each year renewing infection from apparently hibernated basidiospores or conidia. Thus, it is likely that the ancestral species of E. darwinii could have been a circumscribed species that changed its life-strategy during evolution. The geographical origin of this ancestral species is, however, difficult to indicate as the sequences of many species of Exobasidium on North American, South American and South-east Asian Ericaceae are still not available. It is evident that more intensive sampling and the analysis of Exobasidium species from these geographical regions are necessary to determine where and on which host plants the closest relatives of Exobasidium darwinii actually occur.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Begerow D, Bauer R, Boekhout T (2000) Phylogenetic placements of ustilaginomycetous anamorphs as deduced from nuclear LSU rDNA sequences. Mycol Res 104:53–60
Begerow D, Bauer R, Oberwinkler F (1998) Phylogenetic studies on nuclear LSU rDNA sequences of smut fungi and related taxa. Can J Bot 75:2045–2056
Begerow D, Bauer R, Oberwinkler F (2001) Muribasidiospora: Microstromatales or Exobasidiales? Mycol Res 105:798–810
Begerow D, Bauer R, Oberwinkler F (2002) The Exobasidiales: an evolutionary hypothesis. Mycol Prog 1:187–199
Blanz P, Döring H (1995) Taxonomic relationships in the genus Exobasidium (Basidiomycetes) based on ribosomal DNA analysis. Stud Mycol 38:119–128
Blanz P, Oberwinkler F (1983) A contribution to the species definition in the genus Exobasidium (Basidiomycetes). Syst Appl Microbiol 4:199–206
Boekhout T, Fell JW, O’Donnell K (1995) Molecular systematics of some yeast-like anamorphs belonging to the Ustilaginales and Tilletiales. Stud Mycol 38:175–183
Burt EA (1915) The Thelephoraceae of North America 4. Exobasidium. Ann M Bot Gard 2:627–658
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Gardner DE (1985) Red leaf disease of native Hawaiian Vaccinium species caused by Exobasidium vaccinii. Plant Dis 89:805–806
Gatesy J, DeSalle R, Wheeler W (1993) Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Mol Phylogenet Evol 2:152–157
Giribet G, Wheeler WC (1999) On gaps. Mol Phylogenet Evol 13:132–143
Gómez LD, Kisimova-Horovitz L (1998a) (1997) Basidiomicetos de Costa Rica: Exobasidiales, Cryptobasidiales. Notas históricas, taxonómicas y fitogeográficas. Rev Biol Trop 45:1293–1310
Gómez LD, Kisimova-Horovitz L (1998b) Basidiomicetos de Costa Rica: Nuevas especies de Exobasidium (Exobasidiaceae) y registros de Cryptobasidiales. Rev Biol Trop 46:1081–1093
Hotson JW (1927) A new species of Exobasidium. Phytopathology 17:207–217
Huelsenbeck JP, Rannala B (2004) Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst Biol 53:904–913
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinforma Appl Note 17:754–755
Huelsenbeck JP, Larget B, Miller RE, Ronquist F (2002) Potential applications and pitfalls of Bayesian inference of phylogeny. Syst Biol 51:673–688
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program (outlines version 6). Brief Bioinform 9:286–298
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Lutz M, Bauer R, Begerow D, Oberwinkler F, Triebel D (2004) Tuberculina: rust relatives attack rusts. Mycologia 96:614–626
Lutz M, Vánky K, Bauer R (2011) Melanoxa, a new genus in the Urocystidales (Ustilaginomycotina). Mycol Prog. doi:10.1007/s11557-010-0737-7
Nagao H, Ezuka A, Harada Y, Sato T, Kakishima M (2006) Two new species of Exobasidium causing Exobasidium diseases on Vaccinium spp. in Japan. Mycoscience 47:277–283
Nagao H, Ezuka A, Ohkubo H, Kakishima M (2001) A new species of Exobasidium causing witches’ broom on Rhododendron wadanum. Mycoscience 42:549–554
Nannfeldt JA (1981) Exobasidium, a taxonomic reassessment applied to the European species. Symb Bot Ups 23(2):1–72
Nickerson NL (1984) A previously unreported disease of cranberries caused by Exobasidium perenne sp. nov. Can J Plant Pathol 6:218–220
O’Donnell KL (1992) Ribosomal DNA internal transcribed spacers are highly divergent in the phytopathogenic ascomycete Fusarium sambucinum (Gibberella pulicaris). Curr Genet 22:213–220
O’Donnell KL (1993) Fusarium and its near relatives. In: Reynolds DR, Taylor JW (eds) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, pp 225–233
Piepenbring M, Espinoza J, Saldaña L, Cáceres O (2010) New records, host plants, morphological and molecular data of Exobasidiales (Basidiomycota) from Panama. Nova Hedwig 91:231–242
Posada D, Crandall KA (1998) MODELTEST, testing the model of DNA substitution. Bioinformatics 14:817–818
Powell EA, Kron KA (2002) Hawaiian blueberries and their relatives – A phylogenetic analysis of Vaccinium sections Macropelma, Myrtillus, and Hemimyrtillus (Ericaceae). Syst Bot 27:768–779
Raciborski M (1909) Parasitische und epiphytische Pilze Java’s. Bull Int Acad Sci Cracovie Cl Sci Math 3:346–394
Ramakrishnan TS, Ramakrishnan K (1949) Exobasidium from South India. Proc Indian Acad Sci B 29:5–12
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sakai AK, Wagner WL, Mehrhoff LA (2002) Patterns of endangerment in the Hawaiian flora. Syst Biol 51:276–302
Savile DBO (1959) Notes on Exobasidium. Can J Bot 37:641–656
Sundström KR (1964) Studies of the physiology, morphology and serology of Exobasidium. Symb Bot Ups 18(3):1–89
Vander Kloet SP (1993) Biosystematic studies of Vaccinium section Macropelma (Ericaceae) in Hawaii. Pacific Sci 47:76–85
Vander Kloet SP (1996) Taxonomy of Vaccinium sect. Macropelma (Ericaceae). Syst Bot 21:355–364
Vander Kloet SP, Dickinson TA (2009) A subgeneric classification of the genus Vaccinium and the metamorphosis of V. section Bracteata Nakai: more terrestrial and less epiphytic in habit, more continental and less insular in distribution. J Plant Res 122:253–268
Wagner WL, Herbst DR, Sohmer SH (1990) Manual of the flowering plants of Hawai‘i. Bishop Museum Special Publication 83. University of Hawaii Press, Bishop Museum Press, Honolulu
Acknowledgements
We thank the anonymous reviewers for their helpful comments on the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Taxonomical novelty: Exobasidium darwinii M. Piątek & M. Lutz
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Piątek, M., Lutz, M. & Welton, P. Exobasidium darwinii, a new Hawaiian species infecting endemic Vaccinium reticulatum in Haleakala National Park. Mycol Progress 11, 361–371 (2012). https://doi.org/10.1007/s11557-011-0751-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-011-0751-4