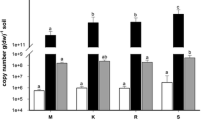
Abstract
We used direct DNA amplification from soil extracts to analyze microbial communities from an elevational transect in the German Alps by parallel metabarcoding of bacteria (16S rRNA), fungi (ITS2), and myxomycetes (18S rRNA). For the three microbial groups, 5710, 6133, and 261 operational taxonomic units (OTU) were found. For the latter group, we can relate OTUs to barcodes from fruit bodies sampled over a 4-year period. The alpha diversity of myxomycetes was positively correlated with that of bacteria. Vegetation type was found to be the main explanatory parameter for the community composition of all three groups and a substantial species turnover with elevation was observed. Bacteria and fungi display similar community responses, driven by symbiont species and plant substrate quality. Myxamoebae show a more patchy distribution, though still clearly stratified between taxa, which seems to be a response to both structural properties of the habitat and interaction with specific bacterial and fungal taxa. Finally, we report a high number of myxomycete OTUs not represented in a reference database from fructifications, which might represent novel species.





Similar content being viewed by others
References
Abarenkov K, Henrik Nilsson R, Larsson K-H, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vrålstad T, Liimatainen K, Peintner U, Kõljalg U (2010) The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytol 186:281–285
Adl MS, Gupta VVSR (2006) Protists in soil ecology and forest nutrient cycling. Can J For Res 36:1805–1817
Adl SM, Simpson AGB, Lane CE et al (2012) The revised classification of eukaryotes. J Eukaryot Microbiol 59:1–45
Anderson MJ (2001) A new method for non parametric multivariate analysis of variance. Austral Ecol 26:32–46
Bahram M, Kohout P, Anslan S, Harend H, Abarenkov K, Tedersoo L (2016) Stochastic distribution of small soil eukaryotes resulting from high dispersal and drift in a local environment. ISME J 10:885–896
Bailey VL, Fansler SJ, Stegen JC, McCue LA (2013) Linking microbial community structure to β-glucosidic function in soil aggregates. ISME J 7:2044–2053
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
Bjørnlund L, Rønn R (2008) “David and Goliath” of the soil food web – flagellates that kill nematodes. Soil Biol Biochem 40:2032–2039
Bjørnlund L, Mørk S, Vestergård M, Rønn R (2006) Trophic interactions between rhizosphere bacteria and bacterial feeders influenced by phosphate and aphids in barley. Biol Fertil Soils 43:1–11
Boenigk J, Ereshefsky M, Hoef-Emden K, Mallet J, Bass D (2012) Concepts in protistology: species definitions and boundaries. Eur J Protistol 48:96–102
Bonkowski M (2002) Protozoa and plant growth: trophic links and mutualism. Eur J Protistol 37:363–365
Bonkowski M (2004) Protozoa and plant growth: the microbial loop in soil revisited. New Phytol 162:617–631
Borg Dahl M, Priemé A, Brejnrod A et al (2017) Warming, shading and a moth outbreak reduce tundra carbon sink strength dramatically by changing plant cover and soil microbial activity. Sci Rep 7:16035
Borg Dahl M, Brejnrod AD, Unterseher M, Hoppe T, Feng Y, Novozhilov Y, Sørensen SJ, Schnittler M (2018a) Genetic barcoding of dark-spored myxomycetes (Amoebozoa) – identification, evaluation and application of a sequence similarity threshold for species differentiation in NGS studies. Mol Ecol Resour 18:306–318
Borg Dahl M, Shchepin ON, Schunk C et al (2018b) A four year survey reveals a coherent pattern between occurrence of fruit bodies and soil amoebae populations for nivicolous myxomycetes. Sci Rep 8:11662
Brundrett MC (2002) Coevolution of roots and mycorrhizas of land plants. New Phytol 154:275–304
Chagnon PL, Bradley RL, Maherali H, Klironomos JN (2013) A trait-based framework to understand life history of mycorrhizal fungi. Trends Plant Sci 18:484–491
Chase JM, Kraft NJB, Smith KG et al (2011) Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2:art24
Choma M, Bárta J, Šantrůčková H, Urich T (2016) Low abundance of Archaeorhizomycetes among fungi in soil metatranscriptomes. Sci Rep 6:38455
Churchman K (2015) Brock Biology of Microorganisms14th edn. Pearson Higher Education, New York
Crump BC, Amaral-Zettler LA, Kling GW (2012) Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISME J 6:1629–1639
Dagamac NHA, Rojas C, Novozhilov YK et al (2017) Speciation in progress? A phylogeographic study among populations of Hemitrichia serpula (Myxomycetes). PLoS One 12:1–22
Day A (2012) Heatmap.plus: Heatmap with more sensible behavior. R package version 1.3
de Boer W, Folman LB, Summerbell RC et al (2005) Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29:795–811
de Vries FT, Manning P, Tallowin JRB, Mortimer SR, Pilgrim ES, Harrison KA, Hobbs PJ, Quirk H, Shipley B, Cornelissen JHC, Kattge J, Bardgett RD (2012) Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol Lett 15:1230–1239
Deslippe JR, Hartmann M, Simard SW, Mohn WW (2012) Long-term warming alters the composition of Arctic soil microbial communities. FEMS Microbiol Ecol 82:303–315
Edgar R (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Edgar R (2016) SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv. 074161
Edgar R, Flyvbjerg H (2014) Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31:3476–3482
Feng Y, Schnittler M (2015) Sex or no sex? Group I introns and independent marker genes reveal the existence of three sexual but reproductively isolated biospecies in Trichia varia (Myxomycetes). Org Divers Evol 15:631–650
Feng Y, Klahr A, Janik P, Ronikier A, Hoppe T, Novozhilov YK, Schnittler M (2016) What an intron may tell: several sexual biospecies coexist in Meriderma spp. (Myxomycetes). Protist 167:234–253
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Finlay RD (2008) Ecological aspects of mycorrhizal symbiosis: with special emphasis on the functional diversity of interactions involving the extraradical mycelium. J Exp Bot 59:1115–1126
Finlay RD, Read DJ (1986) The structure and function of the vegetative mycelium of ectomycorrhizal plants: I. Translocation of 14C-labelled carbon between plants interconnected by a common mycelium. New Phytol 103:143–156
Fiore-Donno AM, Kamono A, Meyer M, Schnittler M, Fukui M, Cavalier-Smith T (2012) 18S rDNA phylogeny of Lamproderma and allied genera (Stemonitales, myxomycetes, Amoebozoa). PLoS One 7:e35359
Fiore-Donno AM, Clissmann F, Meyer M, Schnittler M, Cavalier-Smith T (2013) Two-gene phylogeny of bright-spored myxomycetes (slime-moulds, superorder Lucisporidia). PLoS One 8:e62586
Fiore-Donno AM, Weinert J, Wubet T, Bonkowski M (2016) Metacommunity analysis of amoeboid protists in grassland soils. Sci Rep 6:19068
Franklin RB, Mills AL (2003) Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol Ecol 44:335–346
Fukasawa Y, Hyodo F, Kawakami S (2018) Foraging association between myxomycetes and fungal communities on coarse woody debris. Soil Biol Biochem 121:95–102
Geisen S, Bonkowski M (2017) Methodological advances to study the diversity of soil protists and their functioning in soil food webs. Appl Soil Ecol. https://doi.org/10.1016/j.apsoil.2017.05.021
Geisen S, Cornelia B, Jörg R et al (2014) Soil water availability strongly alters the community composition of soil protists. Pedobiologia 57:205–213
Geisen S, Laros I, Vizcaíno A, Bonkowski M, de Groot GA (2015a) Not all are free-living: high-throughput DNA metabarcoding reveals a diverse community of protists parasitizing soil metazoa. Mol Ecol 24:4556–4569
Geisen S, Tveit AT, Clark IM, Richter A, Svenning MM, Bonkowski M, Urich T (2015b) Metatranscriptomic census of active protists in soils. ISME J 9:2178–2190
Geisen S, Mitchell EAD, Wilkinson DM, Adl S, Bonkowski M, Brown MW, Fiore-Donno AM, Heger TJ, Jassey VEJ, Krashevska V, Lahr DJG, Marcisz K, Mulot M, Payne R, Singer D, Anderson OR, Charman DJ, Ekelund F, Griffiths BS, Rønn R, Smirnov A, Bass D, Belbahri L, Berney C, Blandenier Q, Chatzinotas A, Clarholm M, Dunthorn M, Feest A, Fernández LD, Foissner W, Fournier B, Gentekaki E, Hájek M, Helder J, Jousset A, Koller R, Kumar S, la Terza A, Lamentowicz M, Mazei Y, Santos SS, Seppey CVW, Spiegel FW, Walochnik J, Winding A, Lara E (2017) Soil protistology rebooted: 30 fundamental questions to start with. Soil Biol Biochem 111:94–103
Glücksman E, Bell T, Griffiths RI, Bass D (2010) Closely related protist strains have different grazing impacts on natural bacterial communities. Environ Microbiol 12:3105–3113
Goodfellow M, Williams ST (1983) Ecology of actinomycetes. Annu Rev Microbiol 37:189–216
Goslee SC, Urban DL (2007) The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22:1–19
Harris JK, Sahl JW, Castoe TA, Wagner BD, Pollock DD, Spear JR (2010) Comparison of normalization methods for construction of large, multiplex amplicon pools for next-generation sequencing. Appl Environ Microbiol 76:3863–3868
Hess S, Sausen N, Melkonian M (2012) Shedding light on vampires: the phylogeny of vampyrellid amoebae revisited. PLoS One 7:e31165
Högberg P, Read DJ (2006) Towards a more plant physiological perspective on soil ecology. Trends Ecol Evol 21:548–554
Högberg P, Nordgren A, Buchmann N, Taylor AFS, Ekblad A, Högberg MN, Nyberg G, Ottosson-Löfvenius M, Read DJ (2001) Large-scale forest girdling shows that current photosynthesis drives soil respiration. Nature 411:789–792
Hünninghaus M, Koller R, Kramer S, Marhan S, Kandeler E, Bonkowski M (2017) Changes in bacterial community composition and soil respiration indicate rapid successions of protist grazers during mineralization of maize crop residues. Pedobiologia 62:1–8
Iriberri J, Azúa I, Labirua-Iturburu A, Artolozaga I, Barcina I (1994) Differential elimination of enteric bacteria by protists in a freshwater system. J Appl Bacteriol 77:476–483
Jacquiod S, Stenbæk J, Santos SS, Winding A, Sørensen SJ, Priemé A (2016) Metagenomes provide valuable comparative information on soil microeukaryotes. Res Microbiol 167:436–450
Kamono A, Kojima H, Matsumoto J, Kawamura K, Fukui M (2009) Airborne myxomycete spores: detection using molecular techniques. Naturwissenschaften 96:147–151
Kamono A, Meyer M, Cavalier-Smith T et al (2012) Exploring slime mould diversity in high-altitude forests and grasslands by environmental RNA analysis. FEMS Microbiol Ecol 84:98–109
Katoh K, Misawa K, Kuma K et al (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
King AJ, Freeman KR, McCormick KF et al (2010) Biogeography and habitat modelling of high-alpine bacteria. Nat Commun 1:1–6
Kohler A, Kuo A, Nagy LG et al (2015) Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet 47:410–415
Kowalski DT (1975) The myxomycete taxa described by Charles Meylan. Mycologia 67:448–494
Krashevska V, Bonkowski M, Maraun M, Ruess L, Kandeler E, Scheu S (2008) Microorganisms as driving factors for the community structure of testate amoebae along an altitudinal transect in tropical mountain rain forests. Soil Biol Biochem 40:2427–2433
Lado C (2004) Nivicolous myxomycetes of the Iberian Peninsula: considerations on species richness and ecological requirements. Syst Geogr Plants 74:143–157
Lado C (2005-2019). An on line nomenclatural information system of Eumycetozoa. www.nomen.eumycetozoa.com. Accessed Nov 2017
Lanzén A, Epelde L, Blanco F, Martín I, Artetxe U, Garbisu C (2016) Multi-targeted metagenetic analysis of the influence of climate and environmental parameters on soil microbial communities along an elevational gradient. Sci Rep 6:28257
Lazzaro A, Hilfiker D, Zeyer J (2015) Structures of microbial communities in alpine soils: seasonal and elevational effects. Front Microbiol 6:1–13
Liu QS, Yan SZ, Chen SL (2015) Species diversity of myxomycetes associated with different terrestrial ecosystems, substrata (microhabitats) and environmental factors. Mycol Prog 14:27
Margesin R, Jud M, Tscherko D, Schinner F (2009) Microbial communities and activities in alpine and subalpine soils. FEMS Microbiol Ecol 67:208–218
Martiny JBH, Jones SE, Lennon JT, Martiny AC (2015) Microbiomes in light of traits: a phylogenetic perspective. Science 350:aac9323. https://doi.org/10.1126/science.aac9323
McMurdie JP, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217
Metzing D, Garve E, Matzke-Hajek G et al (2018) Rote Liste und Gesamtartenliste der Farn- und Blütenpflanzen (Tracheophyta) Deutschlands. Natursch Biol Vielfalt 70:313–358
Mueller RC, Paula FS, Mirza BS, Rodrigues JLM, Nüsslein K, Bohannan BJM (2014) Links between plant and fungal communities across a deforestation chronosequence in the Amazon rainforest. ISME J 8:1548–1550
Murase J, Noll M, Frenzel P (2006) Impact of protists on the activity and structure of the bacterial community in a rice field soil. Appl Environ Microbiol 72:5436–5444
Naether A, Foesel BU, Naegele V, Wüst PK, Weinert J, Bonkowski M, Alt F, Oelmann Y, Polle A, Lohaus G, Gockel S, Hemp A, Kalko EKV, Linsenmair KE, Pfeiffer S, Renner S, Schöning I, Weisser WW, Wells K, Fischer M, Overmann J, Friedrich MW (2012) Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl Environ Microbiol 78:7398–7406
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248
Novozhilov YK, Schnittler M, Erastova DA, Okun MV, Schepin ON, Heinrich E (2013) Diversity of nivicolous myxomycetes of the Teberda State Biosphere Reserve (Northwestern Caucasus, Russia). Fungal Divers 59:109–130
Novozhilov YK, Rollins AW, Schnittler M (2017) Ecology and distribution of myxomycetes. In: Stephenson SL, Rojas C (eds) Myxomycetes – biology, systematics, biogeography and ecology. Elsevier, Academic, pp 253–298
Oksanen J, Guillaume FB, Friendly M et al. (2013) R package vegan: ecological diversity. R package version 2.4-2
Osono T, Takeda H (2002) Comparison of litter decomposing ability among diverse fungi in a cool temperate deciduous forest in Japan. Mycologia 94:421–427
Pawlowski J, Burki F (2009) Untangling the phylogeny of amoeboid protists. J Eukaryot Microbiol 56:16–25
Peay KG, Baraloto C, Fine PVA (2013) Strong coupling of plant and fungal community structure across western Amazonian rainforests. ISME J 7:1852–1861
Philippot L, Bru D, Saby NPA, Čuhel J, Arrouays D, Šimek M, Hallin S (2009) Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ Microbiol 11:3096–3104
Portillo MC, Leff JW, Lauber CL et al. (2013) Cell size distributions of soil bacterial and archaeal taxa. Appl Environ Microbiol 79:7610–7617
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Rahanandeh H, Khodakaramian G, Hassanzadeh N et al (2013) Evaluation of antagonistic Pseudomonas against root lesion nematode of tea. Int J Biosci 3:32–40
R Core Team (2013) R: A language and environment for statistical computing. R Found Stat Comput
Rixen C, Stoeckli V, Ammann W (2003) Does artificial snow production affect soil and vegetation of ski pistes? A review. Perspect Plant Ecol Evol Syst 5:219–230
Robeson MS, King AJ, Freeman KR, Birky CW, Martin AP, Schmidt SK (2011) Soil rotifer communities are extremely diverse globally but spatially autocorrelated locally. PNAS 108:4406–4410
Rohlfs M, Albert M, Keller NP, Kempken F (2007) Secondary chemicals protect mould from fungivory. Biol Lett 3:523–525
Ronikier A, Ronikier M (2009) How “alpine” are nivicolous myxomycetes? A worldwide assessment of altitudinal distribution. Mycologia 101:1–16
Rønn R, Mccaig AE, Griffiths BS et al (2002) Impact of protozoan grazing on bacterial community structure in soil microcosms. Appl Environ Microbiol 68:6094–6105
Rosenberg K, Bertaux J, Krome K, Hartmann A, Scheu S, Bonkowski M (2009) Soil amoebae rapidly change bacterial community composition in the rhizosphere of Arabidopsis thaliana. ISME J 3:675–684
Rosling A, Cox F, Cruz-Martinez K, Ihrmark K, Grelet GA, Lindahl BD, Menkis A, James TY (2011) Archaeorhizomycetes: unearthing an ancient class of ubiquitous soil fungi. Science 333:876–879
Roux-Fouillet P, Wipf S, Rixen C (2011) Long-term impacts of ski piste management on alpine vegetation and soils. J Appl Ecol 48:906–915
Ruggiero MA, Gordon DP, Orrell TM et al (2015) A higher level classification of all living organisms. PLoS One 10:1–60
Russel J (2016) MicEco: various functions for analysis for microbial community data. Available from: www.github.com/Russel88/MicEco
Schadt CW, Martin AP, Lipson DA et al (2003) Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301:1359–1361
Schmidt SK, Lipson DA (2004) Microbial growth under the snow: implications for nutrient and allelochemical availability in temperate soils. Plant Soil 259:1–7
Schnittler M, Tesmer JA (2008) A habitat colonisation model for spore-dispersed organisms – does it work with eumycetozoans? Mycol Res 112:697–707
Schnittler M, Novozhilov YK, Romeralo M et al (2012) Myxomycetes and myxomycete-like organisms. In: Frey W (ed) Englers syllabus of plant families, vol 4. Bornträger, Stuttgart, pp 40–88
Schnittler M, Erastova DA, Shchepin ON, Heinrich E, Novozhilov YK (2015) Four years in the Caucasus – observations on the ecology of nivicolous myxomycetes. Fungal Ecol 14:105–115
Shang Y, Sikorski J, Bonkowski M et al (2017) Inferring interactions in complex microbial communities from nucleotide sequence data and environmental parameters. PLoS One 12:e173765
Shchepin O, Novozhilov Y, Schnittler M (2014) Protistology Nivicolous myxomycetes in agar culture: some results and open problems. Potistology 8:53–61
Shchepin ON, Novozhilov YK, Schnittler M (2016) Disentangling the taxonomic structure of the Lepidoderma chailletii-carestianum species complex (Myxogastria, Amoebozoa): genetic and morphological aspects. Protistology 10:120–129
Shchepin ON, Schnittler M, Erastva DA et al (2019) Community of dark-spored myxomycetes in ground litter and soil of taiga forest (Nizhne-Svirskiy Reserve, Russia) revealed by DNA metabarcoding. Fungal Ecol 39:80–93. https://doi.org/10.1016/j.funeco.2018.11.006
Stephenson SL, Feest A (2012) Ecology of soil eumycetozoans. Acta Protozool 51:201–208
Stephenson SL, Schnittler M (2017) Myxomycetes. In: Archibald JM, Simpson AGB, Slamovits CH et al (eds) Handbook of the Protists. Springer, Berlin, pp 1405–1432
Stephenson SL, Novozhilov YK, Schnittler M (2000) Distribution and ecology of myxomycetes in high-latitude regions of the northern hemisphere. J Biogeogr 27:741–754
Stephenson SL, Schnittler M, Novozhilov YK (2008) Myxomycete diversity and distribution from the fossil record to the present. Biodivers Conserv 17:285–301
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tansley IB (1994) Review no. 62. The phytosociology of myxomycetes. New Phytol 126:175–201
Taylor AFS, Alexander I (2005) The ectomycorrhizal symbiosis: life in the real world. Mycologist 19:102–112
Taylor WD, Berger J (1976) Growth responses of cohabiting ciliate protozoa to various prey bacteria. Can J Zool 54:1111–1114
Taylor KM, Feest A, Stephenson SL (2015) The occurrence of myxomycetes in wood. Fungal Ecol 17:179–182
Tedersoo L, Pärtel K, Jairus T, Gates G, Põldmaa K, Tamm H (2009) Ascomycetes associated with ectomycorrhizas: molecular diversity and ecology with particular reference to the Helotiales. Environ Microbiol 11:3166–3178
Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Poldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Partel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson KH, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo LD, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, de Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K (2014) Global diversity and geography of soil fungi. Science 346:1256688–1256689
Tiunov AV, Semenina EE, Aleksandrova AV et al (2015) Stable isotope composition (δ13 C and δ15 N values) of slime molds: placing bacterivorous soil protozoans in the food web context. Rapid Commun Mass Spectrom 29:1465–1472
Treseder KK, Kivlin SN, Hawkes CV (2011) Evolutionary trade-offs among decomposers determine responses to nitrogen enrichment. Ecol Lett 14:933–938
Urich T, Lanzén A, Qi J, Huson DH, Schleper C, Schuster SC (2008) Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527
van der Heijden MGA, Bardgett RD, van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310
van der Putten WH, Bardgett RD, Bever JD, Bezemer TM, Casper BB, Fukami T, Kardol P, Klironomos JN, Kulmatiski A, Schweitzer JA, Suding KN, van de Voorde TFJ, Wardle DA (2013) Plant-soil feedbacks: the past, the present and future challenges. J Ecol 101:265–276
von Mering C, Hugenholtz P, Raes J et al (2007) Quantitative phylogenetic assessment of microbial communities in diverse environments. Science 315:126–130
Wardle DA, Bardgett RD, Klironomos JN, Setälä H, van der Putten W, Wall DH (2004) Ecological linkages between aboveground and belowground biota. Science 304:1629–1633
Whitaker J, Ostle N, Nottingham AT, Ccahuana A, Salinas N, Bardgett RD, Meir P, McNamara N, Austin A (2014) Microbial community composition explains soil respiration responses to changing carbon inputs along an Andes-to-Amazon elevation gradient. J Ecol 102:1058–1071
Wickham H (2010) Ggplot2: elegant graphics for data analysis. Springer-Verlag, New York
Xiong W, Jousset A, Guo S et al (2017) Soil protist communities form a dynamic hub in the soil microbiome. ISME J:1–5
Yashiro E, Pinto-figueroa E, Buri A et al (2016) Local environmental factors drive divergent grassland soil bacterial communities in the western Swiss Alps. Appl Environ Microbiol 82:6303–6316
Zinger L, Lejon DPH, Baptist F, Bouasria A, Aubert S, Geremia RA, Choler P (2011) Contrasting diversity patterns of crenarchaeal, bacterial and fungal soil communities in an alpine landscape. PLoS One 6:e19950
Acknowledgments
MD thanks Dr. Martin Steen Mortensen for assistance with the robot soil extraction and Luma George Odish for laboratorial assistance in the preparation of the Illumina library. Dr. Samuel Jacquiod helped with advice and assistance for the statistical calculations. We also thank Stefan Kellerer and Klaus Kellner, the wardens of the Tröglhütte of the DAV (Deutscher Alpenverein), for making our stay at the field site pleasant. We wish to express general gratitude to researchers for sharing their data and programming codes as well as engaging in online discussion forums (particularly Stack Overflow), which has benefited the analysis in this study.
Funding
Funding for this study was provided in the frame of a Ph.D. position for MD within the Research Training Group RESPONSE (RTG 2010), supported by the Deutsche Forschungsgemeinschaft (DFG).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
ESM 1
List of primers used to amplify the three target communities. (XLSX 9 kb)
ESM 2
DNA reads of mock samples. (XLSX 32 kb)
ESM 3
Sample quality: Illumina quality statistics, OTU accumulation curves, and overview of significant relations between the numbers of DNA reads and the numbers of OTUs. (PDF 2149 kb)
ESM 4
OTU tables. (XLSX 2472 kb)
ESM 5
Maximum-likelihood phylogenetic tree of the most abundant myxomycete OTUs. (PDF 1432 kb)
ESM 6
Analysis script. (PDF 98 kb)
ESM 7
Result script. (PDF 115 kb)
ESM 8
Functional classification. (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Borg Dahl, M., Brejnrod, A.D., Russel, J. et al. Different Degrees of Niche Differentiation for Bacteria, Fungi, and Myxomycetes Within an Elevational Transect in the German Alps. Microb Ecol 78, 764–780 (2019). https://doi.org/10.1007/s00248-019-01347-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01347-1